Benchmark (dorado)
1. Introduction
This benchmark compares the computational time for basecalling of nanopore sequencing waveform data using Dorado on hardware equipped with eight NVIDIA H100 SXM 80GB GPUs.
Dorado is a basecaller developed for Oxford Nanopore's DNA sequencers. A basecaller is software that converts waveform data obtained by measuring DNA molecules into base sequences. This transformation employs machine learning models, and GPUs are used as accelerators to rapidly and accurately convert several terabytes of waveform data into base sequences.
2. Verification GPU Environment
To compare the effective performance of the application, performance evaluations were conducted using nodes equipped with both H100 and V100 GPUs. The H100-equipped node used for verification was executed on a bare-metal server provided by Sakura Internet, featuring their high-performance PHY infrastructure (henceforth referred to as high-performance PHY), while the V100-equipped node was executed on the Thin computation node (Type 2b) of the NIG supercomputer system (henceforth referred to as NIG igt). Both environments were utilised as single-node configurations, with no application performance evaluations conducted on multi-node setups.
To avoid performance discrepancies due to differences in GPU drivers and other system configurations, the software environment settings from the NIG igt were adopted as verification conditions, and the same setup was replicated on the high-performance PHY.
The hardware and software environments of the nodes used for the verification are shown in Table 1.
Table 1: Verification Node Configuration
| NIG igt | High-Performance PHY | |
|---|---|---|
| Hardware Configuration | ||
| CPU (Total Cores) | Intel Xeon Gold 6136 3.0GHz x 2 (24 cores) | Intel Xeon Platinum 8480 2.0GHz x 2 (112 cores) |
| Memory | DDR4 384GB | DDR5 2.0TB |
| GPU (FP64) | NVIDIA V100 SXM2 16GB (7.8 TFlops) x 4 | NVIDIA H100 SXM5 80GB (33.5 Tflops) x 8 |
| GPU Interconnect | NVLink Hybrid Cube Mesh | NVSwitch Fabric |
| System Disk | NVMe SSD 1.6TB x 1 | NVMe SSD 960GB x 2 (RAID1 configuration) |
| Data Disk | NVMe SSD 3.2TB x 1 | NVMe SSD 7.68TB x 4 |
| Software Configuration | ||
| OS | Ubuntu Server 22.04 LTS | Ubuntu Server 22.04 LTS |
| GPU Driver | 530.30.02 | 530.30.02 |
| CUDA | 12.1 | 12.1 |
| Fabric Manager | N/A | UP |
| Singularity CE | 4.0.0 | 4.0.0 |
2.1 Hardware Configuration
Regarding GPUs, the NIG igt node uses four V100 SXM2 16GB GPUs (Volta GV100 architecture) interconnected via NVLink Hybrid Cube Mesh. In contrast, the high-performance PHY node uses eight H100 SXM5 80GB GPUs (Hopper GH100 architecture) interconnected through NVSwitch, offering high-speed interconnects between the GPUs.
2.2 Software Configuration
The software stack, including the OS distribution, GPU drivers, and management tools, was configured to match as closely as possible between the NIG igt and high-performance PHY environments. This was done to minimise any discrepancies, apart from those related to the application implementation itself.
Given that NIG igt is a managed cluster, changes to drivers that are closely tied to the OS environment can be complex. Thus, the high-performance PHY, being a bare-metal server with greater flexibility, was adapted to match the NIG igt software configuration.
The software environment was set up on the Singularity container platform, which is widely used in actual field research, ensuring consistency with real-world workflows.
3. Storage Environment
Dorado requires a storage environment capable of accommodating sufficiently large datasets with high-speed read and write capabilities. Both NIG igt and high-performance PHY feature high-speed NVMe SSDs in their local storage, which were used as storage areas for application input and output.
The storage configurations of the NIG igt and high-performance PHY nodes, along with I/O performance measurements from sequential fio commands, are shown in Table 2.
The /tmp directory, which is the measurement target, refers to the NVMe SSD on the system disk. At NIG igt, there was little difference in performance across the /data area. On the other hand, at high-performance PHY, there was a performance difference of approximately seven times.
fio Command
fio --direct=1 --filename=/data/test --rw=readwrite --bs=1m --size=80G
Table 2: Storage Configuration and Sequential I/O Performance
| Measurement Target | READ | WRITE | |
|---|---|---|---|
| NIG igt | /tmp | 533MiB/s | 531MiB/s |
| /data | 607MiB/s | 605MiB/s | |
| high-performance PHY | /tmp | 145MiB/s | 153MiB/s |
| /data | 1052MiB/s | 1111MiB/s |
4. Performance Evaluation
Performance evaluations were conducted on both high-performance PHY and NIG igt using several versions of Dorado, employing Docker images (nanoporetech/dorado) for the software environment.
Dorado provides basecalling functionality as well as epigenetic modification detection. Three calculation modes were evaluated:
- without modification calling: A computational mode that does not determine epigenetic modification states.
- with methylation calling 5mCG (hereafter referred to as with 5mCG calling): A computational mode that considers only the 5mCG epigenetic modification state. -with methylation calling 5mCG_5hmCG (hereafter referred to as with 5mCG/5hmCG calling): A computational mode that considers both the 5mCG and 5hmCG epigenetic modification states.
The input data used for the benchmark were FAST5 files, similar to those used in the basecaller benchmark conducted on Amazon. The files were split into 584 parts, each ranging from approximately 1GB to 1.7GB in size. The total dataset for analysis amounted to 765GB.
4.1 Execution of Dorado
The three different computation modes were tested with different Dorado commands (v0.5.1). The pod5 directory contained files that were downloaded from the aforementioned benchmark and converted from FAST5 format to pod5 format.
without modification calling
dorado basecaller \
dna_r10.4.1_e8.2_400bps_hac@v3.5.2 \
pod5/ \
--verbose | \
samtools view --threads 8 -O BAM -o output-wo.bam
with 5mCG calling
dorado basecaller \
dna_r10.4.1_e8.2_400bps_hac@v3.5.2\
pod5/ \
--modified-bases 5mCG \
--verbose | \
samtools view --threads 8 -O BAM -o output-5mCG.bam
with 5mCG/5hmCG calling
dorado basecaller \
dna_r10.4.1_e8.2_400bps_hac@v4.0.0\
pod5/ \
--modified-bases 5mCG_5hmCG \
--verbose | \
samtools view --threads 8 -O BAM -o output-5mCG-5hmCG.bam
4.2 Analysis Time by Node and Version
The analysis times for each node and Dorado version are shown in Table 3. As a reference, results from the Amazon benchmark are also provided, comparing p4d.24xlarge (A100x8) and p3.8xlarge (V100x4). Dorado v0.2.4 with 5mCG calling failed during analysis due to the GPU not being used and the system running out of memory, so it is marked as N.A. in the table. Dorado v0.2.4 with 5mCG/5hmCG calling was not executed due to the high likelihood that the results would be similar to those for 5mCG calling, and the focus was on obtaining results for other parameters (this is indicated by "-" in the table).
On high-performance PHY, the analysis was approximately 8 to 15 times faster in the without modification mode, and about 4 to 6 times faster in other modes, compared to NIG igt.
On high-performance PHY, the without modification mode showed about 1.3 times faster performance in versions v0.2.4 and v0.3.0 compared to v0.2.4 and v0.3.0 onwards, and about 1.25 times faster performance for with 5mCG calling in versions v0.3.0 and v0.5.0. However, for with 5mCG/5hmCG calling, no significant performance differences were observed between the versions.
For NIG igt, the trend differed, with analysis time increasing after version v0.5.0 for all modes. This change likely reflects the optimisations for GPUs like the H100, which may not have been beneficial for the V100s in the NIG igt system.
When comparing the same versions on high-performance PHY, without modification calling was found to be at least twice as fast as other computation modes.
Changes in GPU usage and CPU usage during analysis for each mode can be seen in Figure 1, Figure 2, and Figure 3, and CPU usage trends are shown in Figure 4, Figure 5, and Figure 6. The differences in GPU and CPU usage suggest that in modes other than without modification calling, the frequent switching of processes between GPU and CPU may be due to the consideration of epigenetic modifications.
In contrast, for NIG igt, no significant differences were observed across the computation modes. This indicates that other factors are likely limiting the performance, and further investigation is needed.
Finally, comparing with the Amazon benchmark results, high-performance PHY completes analysis in about half the time of the p4d.24xlarge in without modification mode. Additionally, NIG igt is approximately 1.2 times faster than the p3.8xlarge, which has the same GPU configuration.
Table 3: Comparison of Analysis Speed by Node and Dorado Version
| GPU | Dorado Version | Analysis Time (minutes) w/o modification calling | Analysis Time (minutes) w/ 5mCG calling | Analysis Time (minutes) w/ 5mCG/5hmCG calling |
|---|---|---|---|---|
| high-performance PHY (H100x8) | v0.2.4 | 24.30 | N.A. | - |
| v0.3.0 | 18.03 | 50.05 | 45.97 | |
| v0.5.0 | 18.66 | 39.99 | 42.74 | |
| v0.5.1 | 15.97 | 42.80 | 42.48 | |
| NIG igt (V100x4) | v0.2.4 | 211.13 | 215.06 | 224.93 |
| v0.3.0 | 211.05 | 214.86 | 221.89 | |
| v0.5.0 | 244.70 | 267.30 | 232.26 | |
| v0.5.1 | 242.08 | 263.06 | 231.39 | |
| Amazon Benchmark p4d.24xlarge (A100x8) | v0.2.4 | 48.00 | 48.00 | 54.00 |
| Amazon Benchmark p3.8xlarge (V100x4) | v0.2.4 | 258.00 | 264.00 | 282.00 |
Figure 1: GPU Usage during Analysis with Dorado v0.5.1 (without modification calling)
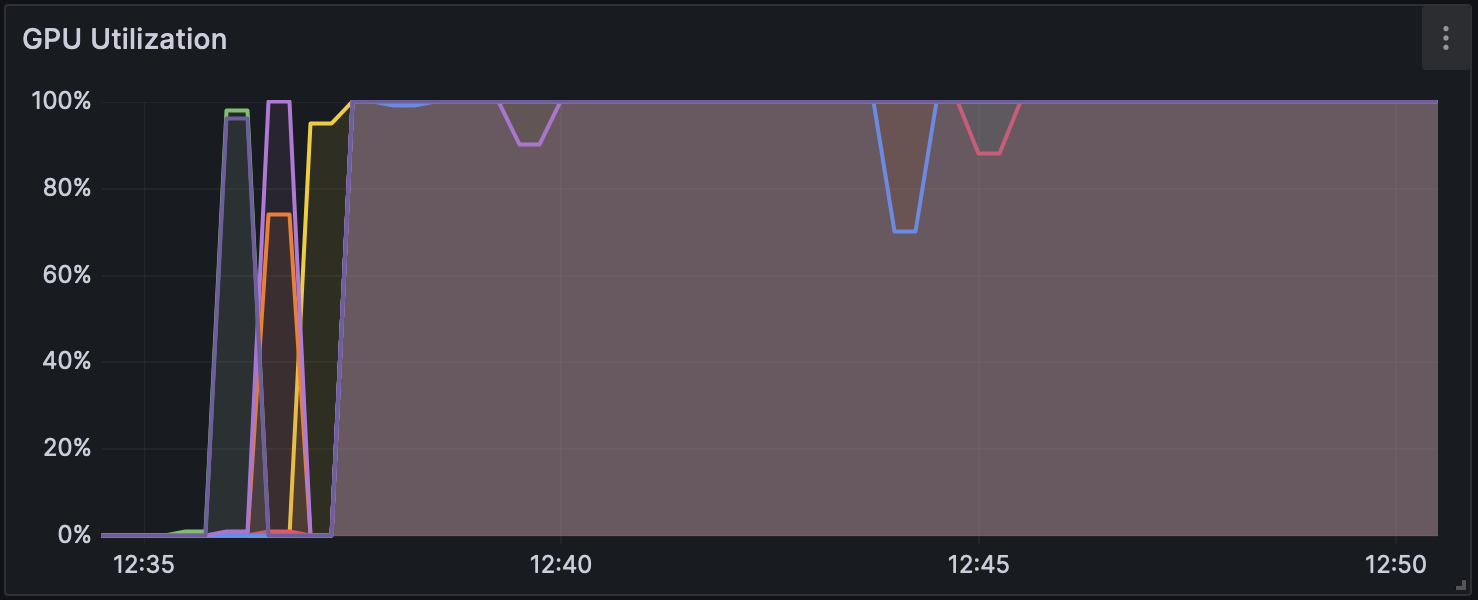
Figure 2: GPU Usage during Analysis with Dorado v0.5.1 (with 5mCG calling)
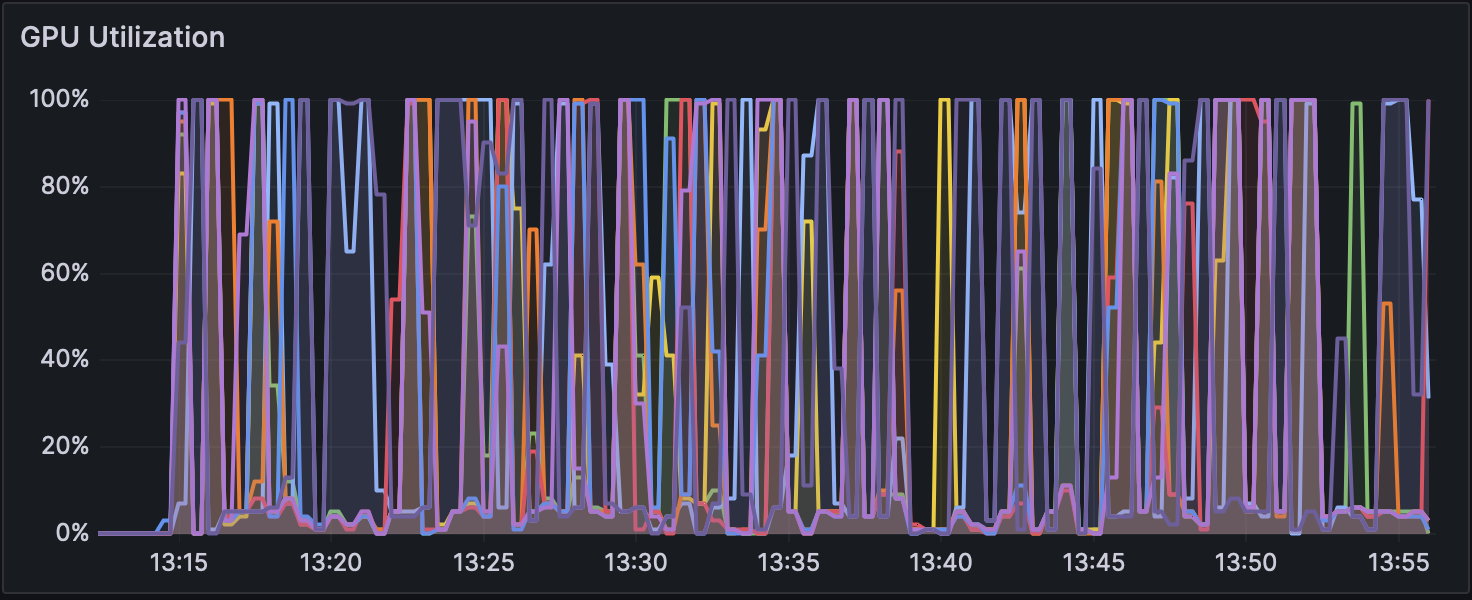
Figure 3: GPU Usage during Analysis with Dorado v0.5.1 (with 5mCG/5hmCG calling)
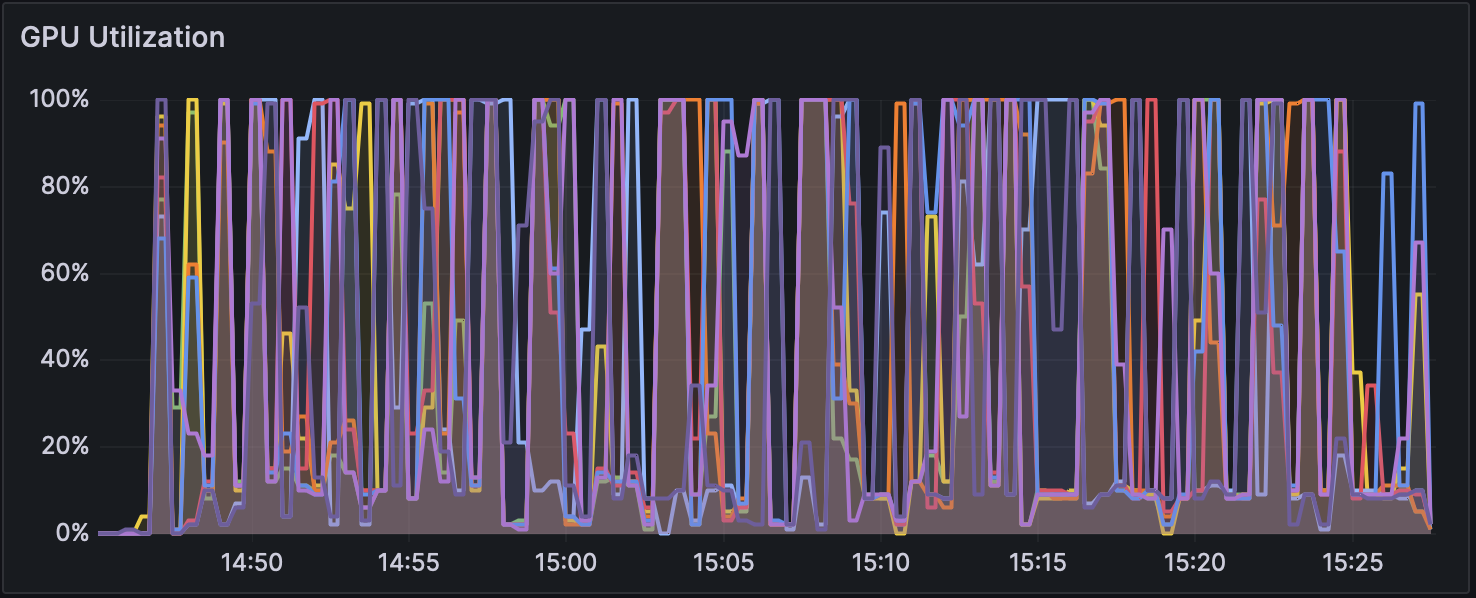
Figure 4: CPU Usage during Analysis with Dorado v0.5.1 (without modification calling)
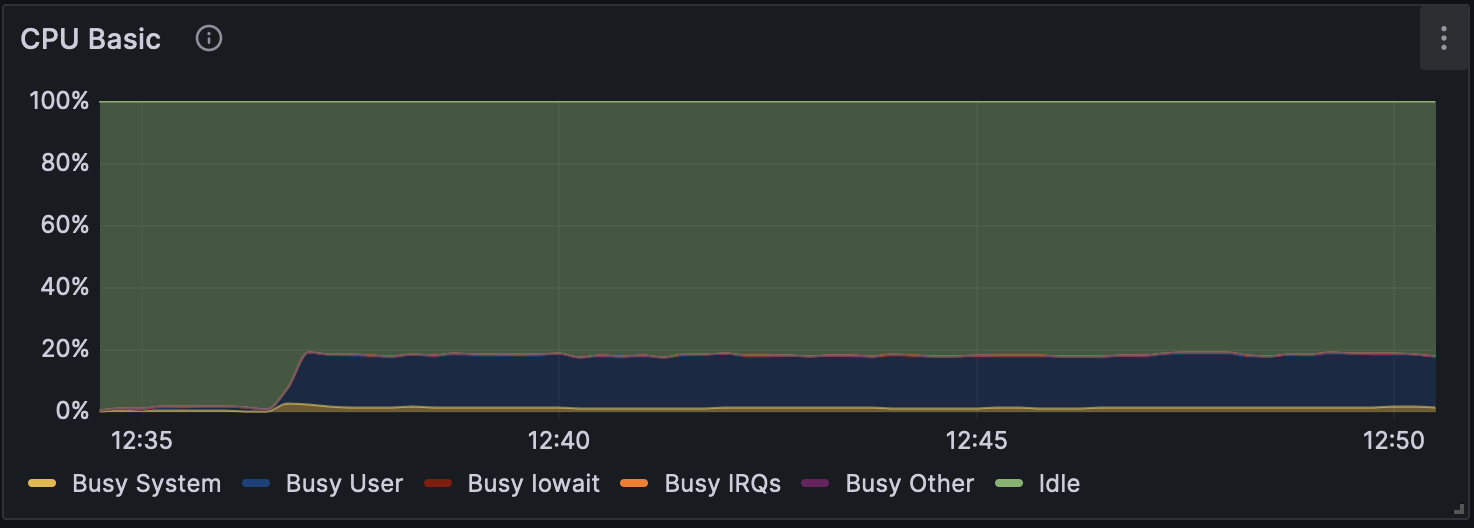
Figure 5: CPU Usage during Analysis with Dorado v0.5.1 (with 5mCG calling)
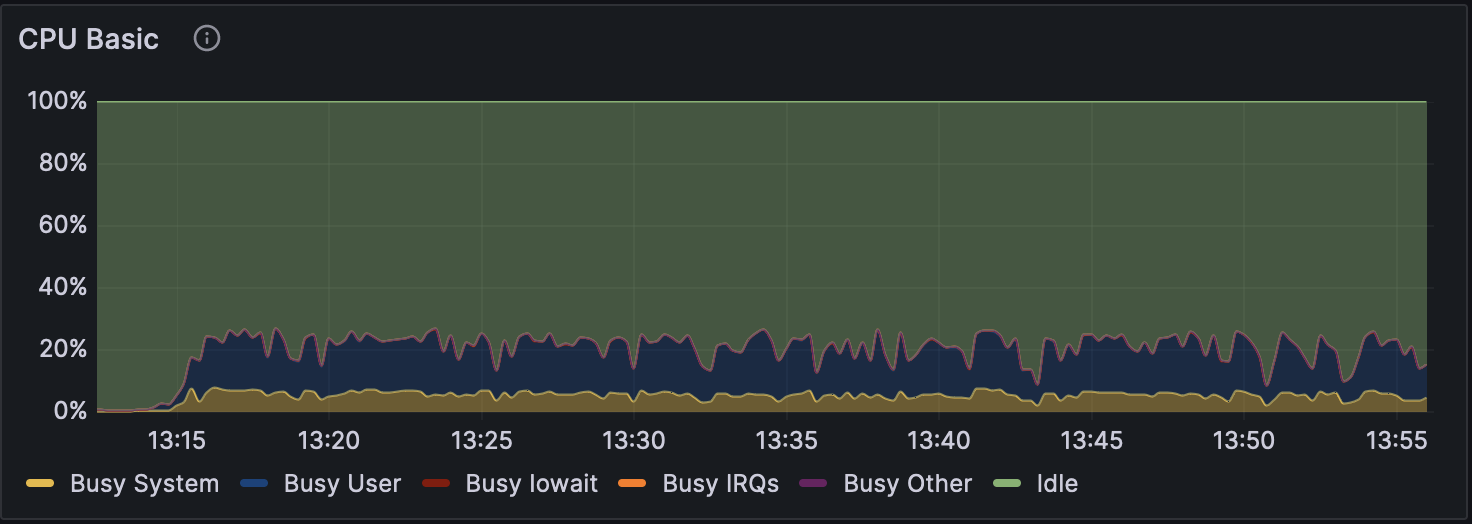
Figure 6: CPU Usage during Analysis with Dorado v0.5.1 (with 5mCG/5hmCG calling)
5. Conclusion
This study demonstrates that the use of computing nodes equipped with the latest H100 GPUs can significantly accelerate genome analysis using Dorado. However, the limited period during which the High-Performance PHY was available left some aspects unresolved, and further investigations are planned in collaboration with relevant parties.
6. About this Article
The findings presented here were generated based on measurements conducted in the fiscal year 2023 as part of the joint research project "Research on the Lifecycle Design of Large-Scale Research Data" (2020–) between the National Institute of Genetics (NIG) DDBJ Centre and Sakura Internet Inc.
7. Article Details and Related Link
Article Creation Date: 6th June 2024
7.1 Project Members
- Tomoya Tanjo1, 2
- Shun Nogawa3
- Kentaro Yamamoto3
- Manabu Ishii3
- Tatsuro Ohta1, 4
- Fumikazu Konishi5
- Tsuyoshi Hachiya3
- Osamu Ogasawara1
1: National Institute of Genetics, Bioinformation and DDBJ Center
2: Joint Support-Center for Data Science Research, BioData Science Initiative
3: Genome Analytics Japan Inc.
4: Chiba University, Institute for Adovanced Academic Research
5: Sakura Internet Inc.

