Hoe to use Slurm GPU node specific queue of Personal genome analysis division
Overview
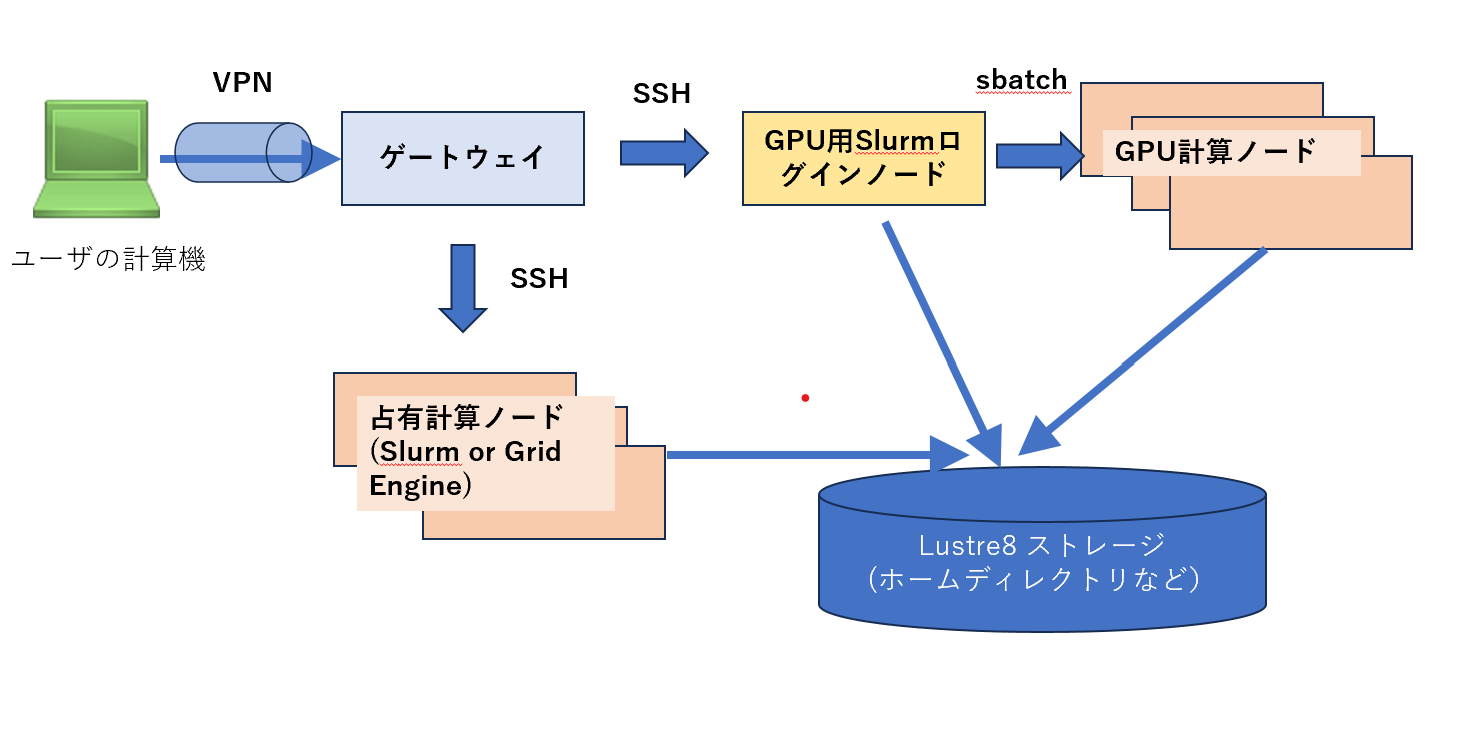
In the Personal Genome Analysis division of the DIG supercomputer, jobs can be submitted to GPU nodes managed under the Slurm resource scheduler.
Ths Slurm cluster assumes that all GPU nodes (4 GPUs) are used to use Parabricks, so another user's job cannot be submitted to the same GPU node at the same time.
Prepare for use
- This service is a billing service, so you should submit a usage plan.
- We will create users home(symbolic) on the Slurm compute node for the GPU, so please fill out the approximate period of your use in the "Purpose of use, etc." of your usage plan or let us know by email.
- If you would like to run Parabrick with rootless docker, fill out this in the "Purpose of use, ets." of your usage plan or let us know by email. (*This is not necessary when using Apptainer (Singularity)).
Usage instructions with Apptainer
In the following procedure, Parabricks v4.0 is executed using the apptainer image file. (For an explanation of Apptainer itself, refer to the Apptainer (Singularity) page)'.
You can use Parabricks image files prepared by the user or under /opt/pkg/nvidia/parabricks deployed on the NIG supercomputer.
Login node for Slurm GPU queue and submit jobs
There is a front server at022vm02 that can submit jobs.
Prerequisite
- You must already be logged in to gwa.ddbj.nig.ac.jp.
Log in to the login node for the Slurm GPU queue.
$ ssh at022vm02
Download sample data. (When executed in /home/$(id -un)/)
$ wget -O parabricks_sample.tar.gz "https://s3.amazonaws.com/parabricks.sample/parabricks_sample.tar.gz"
Unpack the downloaded files and check that the parabricks_sample directory has been created.
$ tar -zxf parabricks_sample.tar.gz
$ ls
................ parabricks_sample ................ ................ ................
Create a job script.
In this procedure, a script named test.sh is created with the following contents.
- Description of the job script
test.sh
#!/bin/bash
#
#SBATCH --partition=all # Select all here
#SBATCH --job-name=test
#SBATCH --output=res.txt
#SBATCH --mem 384000 # Unit MB, allocating 384 GB of total memory for the GPU node
apptainer exec --nv --bind /home/$(id -un):/input_data /opt/pkg/nvidia/parabricks/clara-parabricks_4.0.0-1.sif \
pbrun fq2bam \
--ref /input_data/parabricks_sample/Ref/Homo_sapiens_assembly38.fasta \
--in-fq /input_data/parabricks_sample/Data/sample_1.fq.gz /input_data/parabricks_sample/Data/sample_2.fq.gz \
--out-bam /input_data/parabricks_sample/fq2bam_output.bam
Also, in the partitions of this Slurm cluster (equivalent to queues in AGE), three types are available: igt009, igt016 and all.
$ sinfo -l
Mon Mar 13 10:44:04 2023
PARTITION AVAIL TIMELIMIT JOB_SIZE ROOT OVERSUBS GROUPS NODES STATE NODELIST
igt009 up infinite 1-infinite no NO all 1 reserved igt009
igt016 up infinite 1-infinite no NO all 1 reserved igt016
all* up infinite 1-infinite no NO all 2 reserved igt[009,016]
Submit the job.
$ sbatch test.sh
Check that the job has been submitted.
$ squeue
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
56 all test pg-user PD 0:00 1 (ReqNodeNotAvail, May be reserved for other job)
$
When the job is completed runnning, check the output log and results file.
$ cat res.txt
$ ls parabricks_sample/
Data Ref fq2bam_output.bam fq2bam_output.bam.bai fq2bam_output_chrs.txt
- res.txt … output log
- fq2bam_output.bam, fq2bam_output.bam.bai, fq2bam_output_chrs.txt… results file
Usage instructions with Rootless Docker
This section describes the procedure for using Rootless Docker to analyse by Parabricks.
- Log in to each worker node (GPU node) and start Rootless Docker.
- Log in to the Slurm GPU queue node and submit a job.
Login to each worker node (GPU node) and start Rootless Docker.
Rootless Docker is a Docker that the general account that is not root (administrator account) can also start and run. Normal Docker can only be started and run by root.
Perform this procedure to run Parabricks containers.
Reference: https://docs.nvidia.com/clara/parabricks/4.0.0/GettingStarted.html#gettingstarted
Prerequisite
You must already be logged in to gwa.ddbj.nig.ac.jp. There should be two servers, igt009 and igt016, on which Rootless Docker is started.
Create the directories and files required when starting Rootless Docker in your own home directory on Lustre.
$ mkdir -p /home/$(id -un)/.docker/run_igt009
$ mkdir -p /home/$(id -un)/.docker/run_igt016
$ mkdir -p /home/$(id -un)/.config/docker
$ cat <<EOF > /home/$(id -un)/.config/docker/daemon.json
{"data-root":"/data1/rootless-docker-$(id -un)"}
EOF
The following is the configuration work for the GPU nodes.
Currently, there are two GPUs igt009 and igt016, each of which needs to be configured.
Login to the GPU node
$ ssh <GPU nodename igt009 or igt016>
Create a directory in the NVMe area of the GPU node(/data1) where Parabricks intermediate files etc. are output.
$ mkdir /data1/rootless-docker-$(id -un); chmod 750 /data1/rootless-docker-$(id -un)/;
Start Rootless Docker
$ dockerd-rootless.sh --experimental --storage-driver vfs &
Pull the Parabricks Docker image.
The following example pulls a Docker image of version 4.0.0-1.
$ docker pull nvcr.io/nvidia/clara/clara-parabricks:4.0.0-1
Log out of the GPU node.
$ exit
Submit a job to the login node for the Slurm GPU queue.
A front server at022vm02 is available for submitting jobs.
Prerequisites
- You must have already logged in to gwa.ddbj.nig.ac.jp.
- If you run Parabricks, you must have already started Rootless Docker as described above.
Login to the login node for the Slurm GPU queue.
$ ssh at022vm02
Download sample data. (When executed in /home/$(id -un)/)
$ wget -O parabricks_sample.tar.gz "https://s3.amazonaws.com/parabricks.sample/parabricks_sample.tar.gz"
Extract the downloaded files and Confirm that the parabricks_sample directory has been created.
$ tar -zxf parabricks_sample.tar.gz
$ ls
................ parabricks_sample ................ ................ ................
Create a job script.
In this procedure, create a script named test.sh with the following contents.
※ "source /etc/profile.d/rootless-docker.sh" defines the necessary environment variables and must be filled in.
- Description of the job script
test.sh.
#!/bin/bash
#
#SBATCH --partition=all # Select all here
#SBATCH --job-name=test
#SBATCH --output=res.txt
source /etc/profile.d/rootless-docker.sh
docker run --gpus all --rm --volume /home/$(id -un):/input_data nvcr.io/nvidia/clara/clara-parabricks:4.0.0-1 pbrun fq2bam --ref /input_data/parabricks_sample/Ref/Homo_sapiens_assembly38.fasta --in-fq /input_data/parabricks_sample/Data/sample_1.fq.gz /input_data/parabricks_sample/Data/sample_2.fq.gz --out-bam /input_data/parabricks_sample/fq2bam_output.bam
There are three types of partitions (equivalent to queues in AGE) in this Slurm cluster: igt009, igt016 and all.
$ sinfo -l
Mon Mar 13 10:44:04 2023
PARTITION AVAIL TIMELIMIT JOB_SIZE ROOT OVERSUBS GROUPS NODES STATE NODELIST
igt009 up infinite 1-infinite no NO all 1 reserved igt009
igt016 up infinite 1-infinite no NO all 1 reserved igt016
all* up infinite 1-infinite no NO all 2 reserved igt[009,016]
Submit the job.
$ sbatch test.sh
Confirm that the job has been submitted.
$ squeue
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
56 all test pg-user PD 0:00 1 (ReqNodeNotAvail, May be reserved for other job)
$
When the job execution has completed, confirm the output log and results file.
$ cat res.txt
$ ls parabricks_sample/
Data Ref fq2bam_output.bam fq2bam_output.bam.bai fq2bam_output_chrs.txt
- res.txt is the output log.
- fq2bam_output.bam, fq2bam_output.bam.bai and fq2bam_output_chrs.txt are the result file.