TogoImputation

The Imputation Server is a service to support imputation analysis of SNP array data. 🔗Michigan Imputation Server and 🔗TOPMed Project Imputation Server are public. These servers are located outside Japan and genomic data (SNP array data) had to be uploaded to the servers outside Japan for use.
Therefore, 🔗the Database Center for Life Science has developed the TogoImputation system as a Japanese version of the imputation server that is easy for Japanese researchers to use. This system is currently available in the Personal Genome Analysis division of the NIG supercomputer system.
The imputation workflow used in this server was modified and implemented as a web service with reference to the information (selection of imputation software and setting of parameters) provided by the National Center for Global Health and Medicine in the following AMED project.
Project name: Platform Program for Promotion of Genome Medicine (Research and development research to resolve issues related to international data sharing)
Subject name: "Investigation and practice of ethical and technical issues in genomic medical science research using cloud computing environment"
Introduction
The TogoImputation (beta version) (hereafter referred to as 'this system') is available in the Personal Genome Analysis division of the NIG supercomputer. Researchers(users) can upload their own genomic data to the server and execute the imputation analysis workflow via the web user interface. After the workflow calculations are completed, the imputed genomic data, which are the results of the calculations, can be downloaded. This system can be used securely by using a virtual private network (SSL-VPN) with encrypted communication.
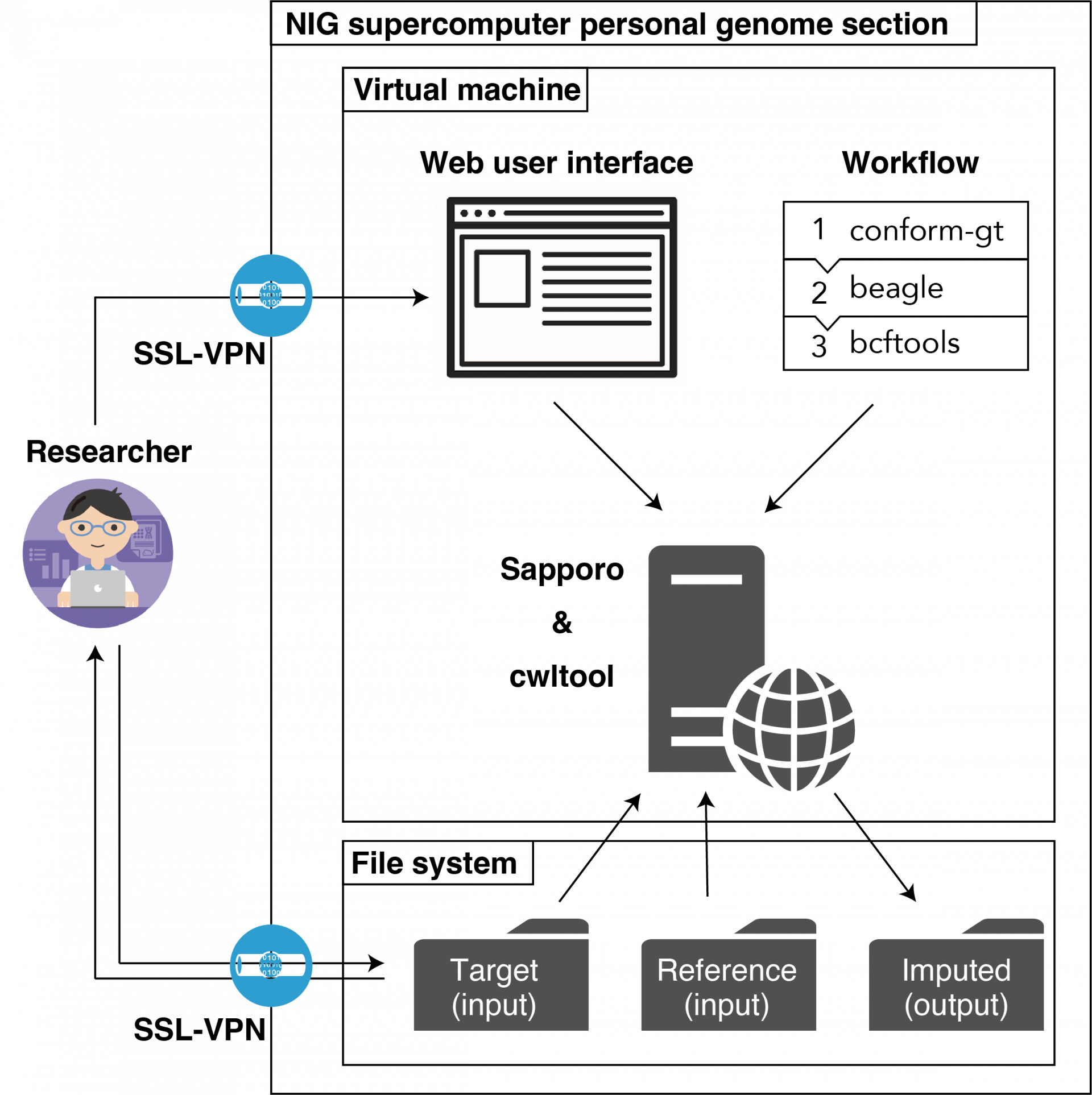
System Diagram of the TogoImputation Researcher illustration were created by TogoTV (©2016 DBCLS TogoTV / CC-BY-4.0).
Available Imputation Algorithms
This system uses the following programmes for imputation analysis.
- Use 🔗conform-gt (version 24May16) to convert the reference / alternative allele of the input SNP array data to match the reference panel data.
- Use 🔗Beagle 5.2 (version 21Apr21.304) for fading and imputation analysis.
- Index the genomic data (VCF file) after imputation using 🔗bcftools (version 1.9).
A series of workflows are implemented in 🔗Common Workflow Language (CWL) and published as an 🔗input-server workflow.
Main input files
The following two datasets are inputs to the imputation workflow.
- Target genotype dataset: A dataset genotyped with SNP microarrays. It is assumed that you upload the data yourself to the server. The file format VCF is supported. (PLINK format is currently not).
- Reference panel dataset: A dataset containing phased haplotypes. Six ready-to-use reference panels are available.
Main input files
- Imputed genotype dataset: Post-imputation genotype dataset. The file format is VCF. The allele dosage data resulting from imputation is output to a DS tag. The estimated allele frequency and imputation quality (dosage R2) are output in the INFO column.
Available reference panel types.
In this system, six types of reference panels are currently available. If you use a reference panel for unrestricted data, it is not necessary to apply to use the data. But for restricted public data, you need to apply to NBDC for data usage of the reference panel data.
| Reference Panel Name | Overview | Access Level | Assembly Version |
|---|---|---|---|
| GRCh37.1KGP | Reference panel '🔗The 1000 Genomes Project'. Includes 2,504 samples belonging to several ancestries | Unrestricted-Access | hg19, GRCh37 |
| GRCh37.1KGP_EAS | Reference panel '🔗The 1000 Genomes Project'. Includes 504 samples belonging to East Asian ancestry | Unrestricted-Access | hg19, GRCh37 |
| GRCh38.1KGP | Reference panel '🔗The 1000 Genomes Project'. Includes 2,548 samples belonging to several ancestries | Unrestricted-Access | GRCh38 |
| GRCh38.1KGP_EAS | Reference panel '🔗The 1000 Genomes Project'. Includes 508 samples belonging to East Asian ancestry | Unrestricted-Access | GRCh38 |
| BBJ1K+GRCh37.1KGP | Reference panel obtained by cross-imputing the reference panel '🔗BioBank Japan project' with the reference panel '🔗The 1000 Genomes Project'. Includes 1,037 samples from the BioBank Japan project and 2,504 samples from several ancestries of the 10,000 Genomes Project (3,541 in total) | Controlled-Access | hg19, GRCh37 |
| BBJ1K+GRCh37.1KGP_EAS | Reference panel obtained by cross-imputing the reference panel '🔗BioBank Japan project' with the reference panel '🔗1000 Genomes Project'. Includes 1,037 samples from the BioBank Japan project and 504 samples belonging to the East Asian ancestry of the 10,000 Genomes Project (1,541 in total) | Controlled-Access | hg19, GRCh37 |
| BBJ1K (GRCh38) | A reference panel obtained by processing the WGS data of the 🔗BioBank Japan project (N=1,026). | Controlled-Access | GRCh38 |
| BBJ2K (GRCh38) | A reference panel obtained by processing the WGS data of the 🔗BioBank Japan project (N=1,964). | Controlled-Access | GRCh38 |
A result comparing the imputation accuracy of different reference panels showed that the restricted public data, BBJ1K+GRCh37.1KGP reference panel, was rated as the most accurate. For more information, see 🔗the paper written about this system.
Notes on using this system to prepare papers.
- Cite the paper in this system.
- Describe the use of the NIG supercomputer in an acknowledgement letter or similar. (Sample text).
- Example text: Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
- If you have used restricted public data, include the accession number of the dataset used in your paper and cite Nucleic Acids Res. 2015, 43 Database issue: D18-D22. In addition, cite the paper in which the dataset was reported (the paper prepared by the data provider of the dataset using the dataset as the basis for the data), or describe an acknowledgement as the following example text.
- Example text: (A part of) The data used for this research was originally obtained by AAAA research project/group led by Prof./Dr. BBBB and available at the website of the NBDC Human Database website of the Database Center for Life Science (DBCLS) / the Joint Support-Center for Data Science Research of the Research Organization of Information and Systems.
How to use the TogoImputation (beta)
This system is for users who use the Personal Genome Analysis division. For information on how to apply for use of it, refer to the Steps from user account application to start of use page in the Personal Genome Analysis division. Users of the personal genome analysis division can use this system according to the following procedure. It is strongly recommended to use a virtual environment with one guacamole per user when using the system.
- Send an application email to the application desk for the use of this system (imputation-server@ddbj.nig.ac.jp)
- The supercomputer administrator will cut out a part of the computer node and start a virtual machine of this system. The remote desktop environment user manual will also be sent to you.
- Log in to the Remote Desktop Environment according to the Remote Desktop Environment User Manual.
- Install Manual to complete the setup.
- Use Tutorial 1 - Using the public reference panel to learn how to use the Imputation Server with publicly available genomic and reference data
- After completing the above steps, you can upload your genomic data (SNP array data), perform an imputation analysis and download the results of the imputation analysis.
- If you use restricted public data, refer to Tutorial 2 - Using the restricted release reference panel.
text:Example of TogoImputation usage application email
I request the use of the TogoImputation (beta version).
I would appreciate it if you could create a new virtual machine environment using guacamole.
Account name for the personal genome analysis division: ________ (e.g. youraccount-pg)
Machine name to start guacamle: ________ (e.g. at001)
Number of cores: ______ (recommended: 16 or more)
RAM: ______ (recommended: 128 GB or more)
Directory to mount: __________ (e.g. /home/ddbjshare-pg [required], /home/youraccount-pg [required])
Please also install singularity.
Applying for data use of restricted public data
Restricted Public Reference Panels are registered in the Japanese Genotype-phenotype Archive (JGA). For how to apply for use of JGA data, see 🔗NBDC Human Database - Data Usage. Refer to the following table for the accession code required when applying.
| Reference Panel Name | Research ID | Dataset ID |
|---|---|---|
| BBJ1K+GRCh37.1KGP | hum0014 | JGAD000679 |
| BBJ1K+GRCh37.1KGP_EAS | hum0014 | JGAD000679 |
| BBJ1K (GRCh38) | hum0014 | JGAD000867 |
| BBJ2K (GRCh38) | hum0014 | JGAD000868 |
Contact us
For any queries regarding this system, contact imputation-server@ddbj.nig.ac.jp.