Tutorial 1 - Using the public reference panel

Procedures for using this system
This system executes workflows in the following steps.
- Prepare test data
- Generate a configuration file for the Imputation Workflow
- Execute the Imputation Workflow
1. Prepare test data
To proceed with the tutorial, download the test data to be used and copy it to the Personal Genome Analysis division of the NIG supercomputer.
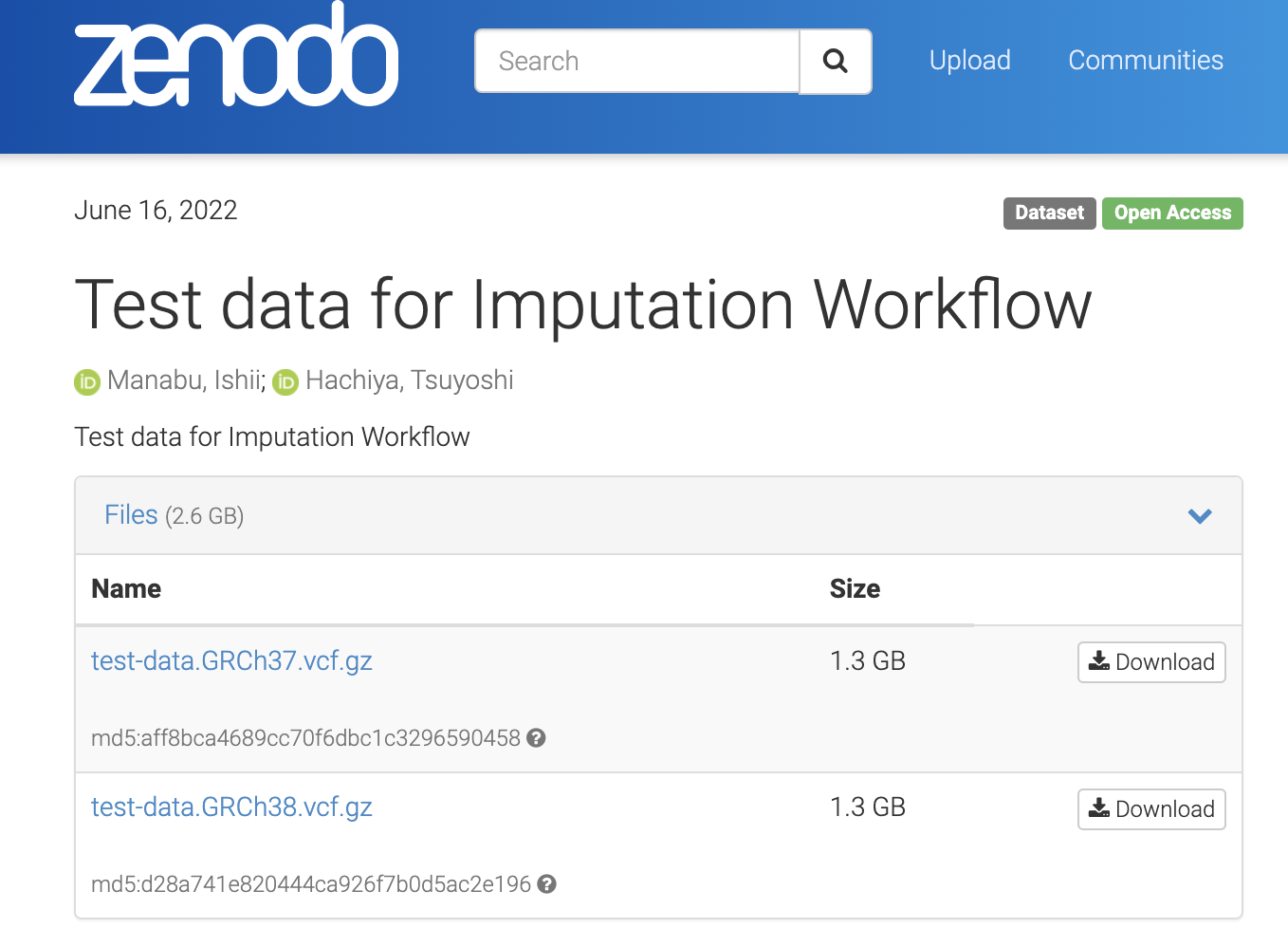
Download the test data
Access 🔗Test data for Imputation Workflow. You can find the following two files.
test-data.GRCh37.vcf.gztest-data.GRCh38.vcf.gz
This time, we will use test-data.GRCh37.vcf.gz, so download it.
Even if you use test-data.GRCh38.vcf.gz, the steps of procedure are the same and you can proceed without any problems if you select GRCh38 if necessary.
Copy it to the Personal Genome Analysis division of the NIG supercomputer
Copy the test data just downloaded.
First, connect the VPN for connecting to the NIG supercomputer.
Next, use the following command to copy the test data that you have just downloaded.
In the following example, the test data you want to copy are in the download folder, and the copy destination is the home directory of your account in the Personal Genome Analysis division of the NIG supercomputer.
scp -i [your private key file] ~/download/test-data.GRCh37.vcf.gz ([your account name])@gwa.ddbj.nig.ac.jp:~/
Test data is now prepared.
2. Generate a configuration file for the Imputation Workflow
Access the following address via guacamole on the NIG supercomputer.
http://localhost:5000
When you actually access it, you will see the following screen.
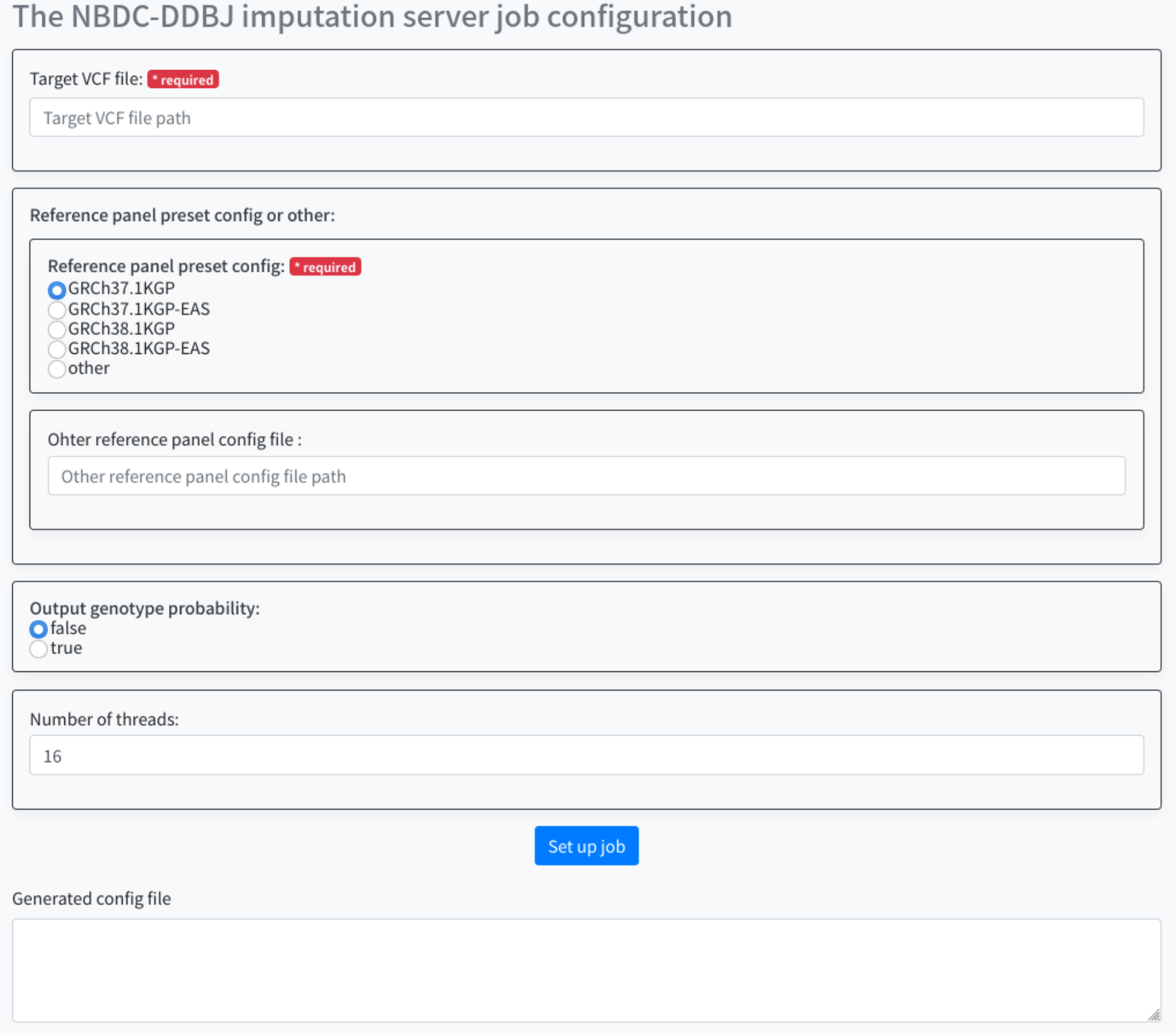
Configure the following items.
- Target VCF file
- Reference panel preset config or other
- Output genotype probability
- Number of threads
For the target VCF file, specify the full path of the VCF file (*.vcf.gz file) to be parsed.
Here, the file that you uploaded is used.
The specific full path will be /home/username/test-data.GRCh37.vcf.gz.
Select the 'Reference panel preset config or'. By default, you can choose for the following four.
- GRCh37.1KGP
- GRCh37.1KGP-EAS
- GRCh38.1KGP
- GRCh38.1KGP-EAS
For more information on each of them, see 🔗Types of Reference Panels available.
If you want to use other than the above as a Reference Panel, select 'other' and specify the one you want to use for the Reference panel config file.
Select 'Output genotyhpe probability'. You can select the following two types. By default, false is selected.
- false
- true
For 'Number of threads', specify the number of threads for the job when running the workflow.
By default, 16 is specified.
After specifying the parameters, press the Set up job button. The generated parameters are displayed at the bottom of the screen. Use this in sapporo-web.
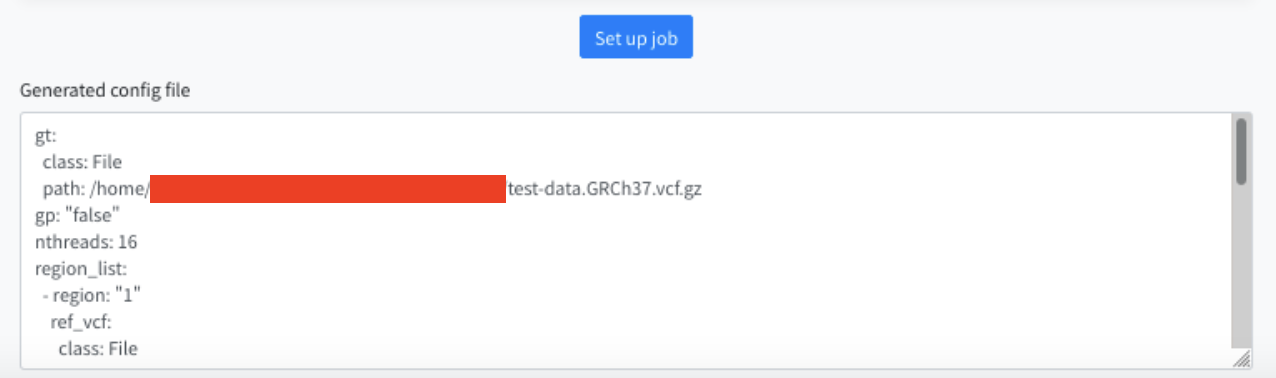
The red filled area is your account name.
3. Execute the Imputation Workflow
Via guacamole, access the following address.
http://localhost:1121
When accessed, the following screen is displayed.
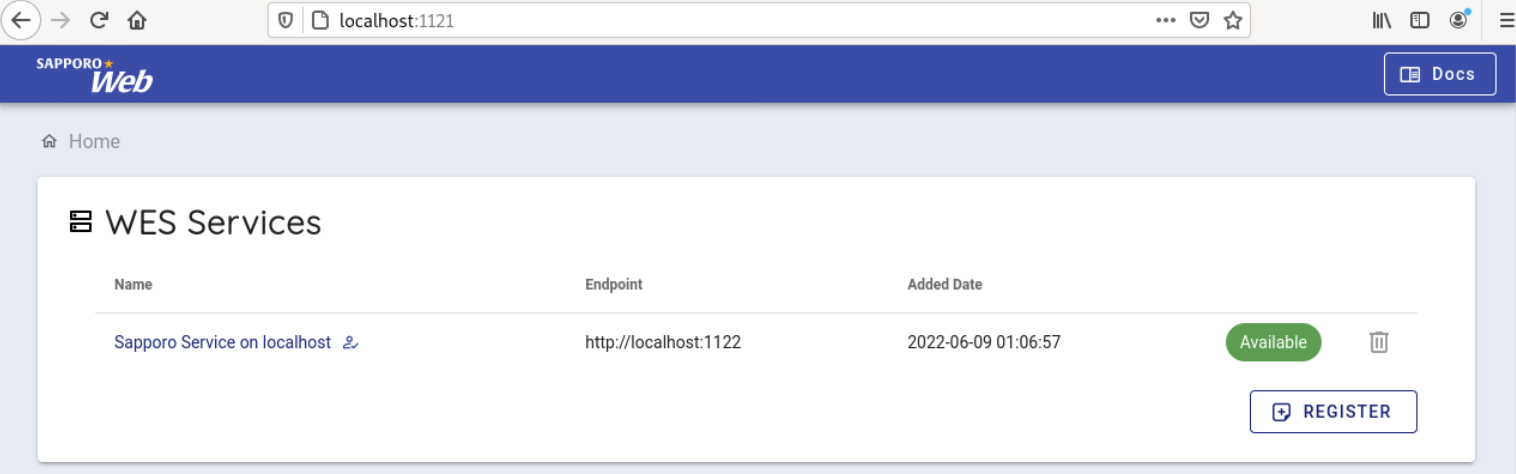
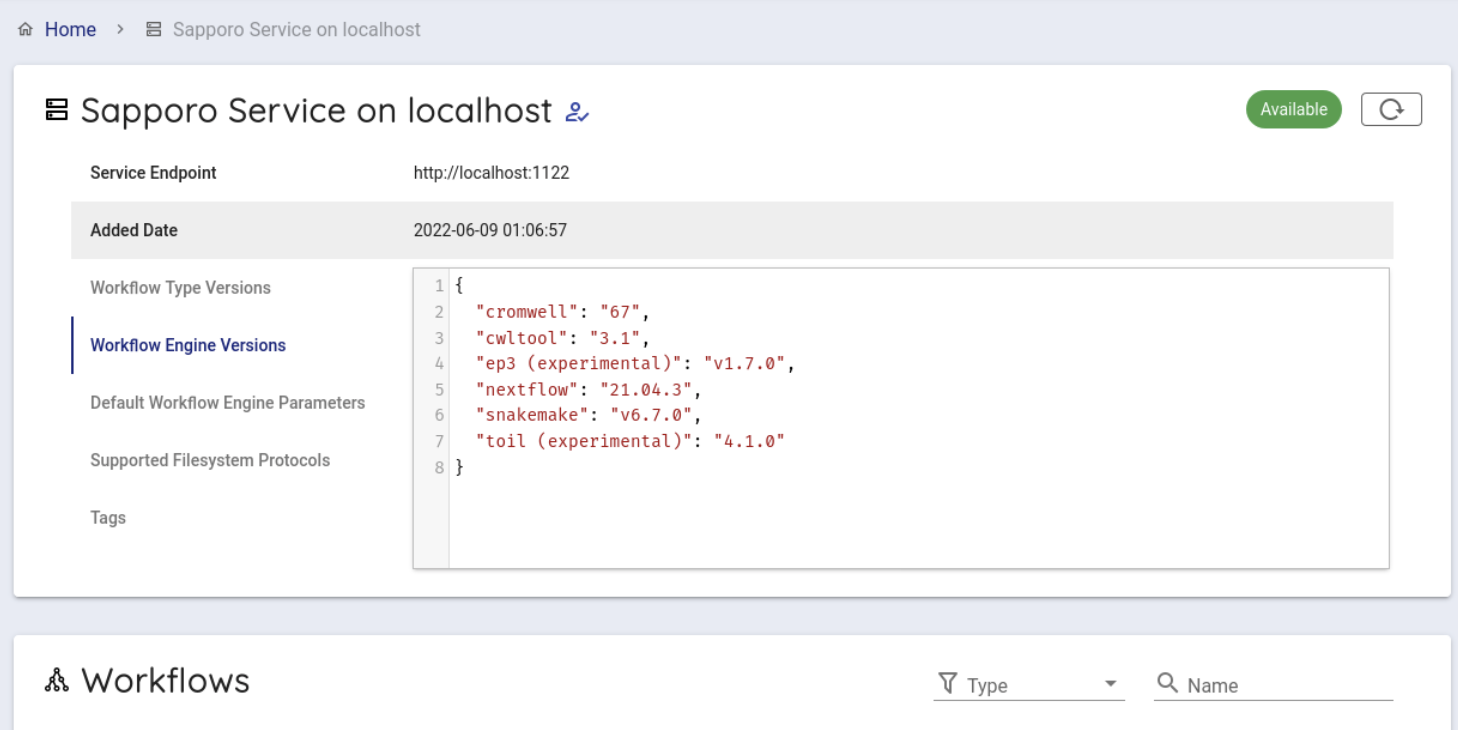
Select 'Sapporo Service on localhost', which is available by default.
When clicked, you can see the following screen.
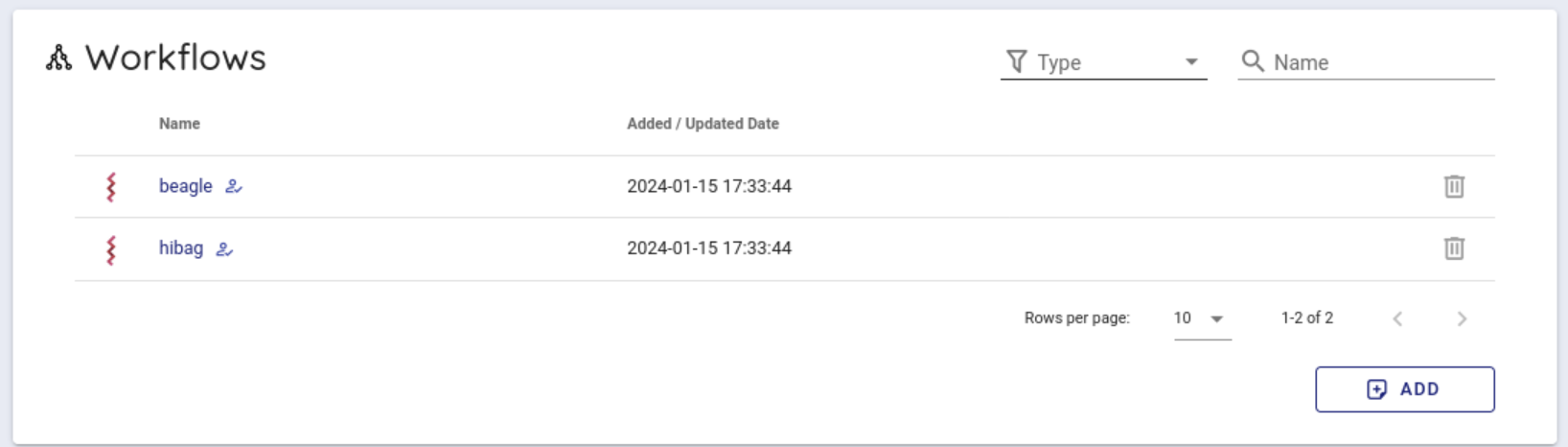
Scroll down a little to use the backend workflows and select 'beagle' from the Workflows item and click it.
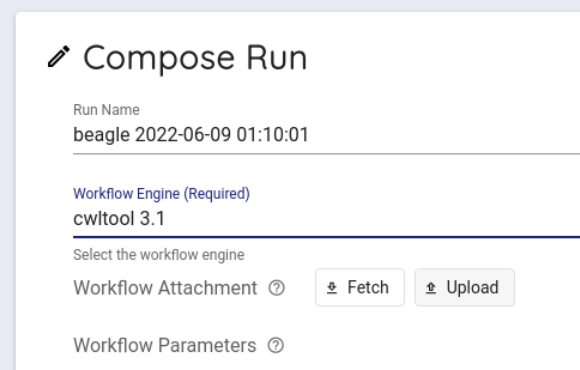
Select cwltool 3.1 from the Workflow Engine item of Compose Run.
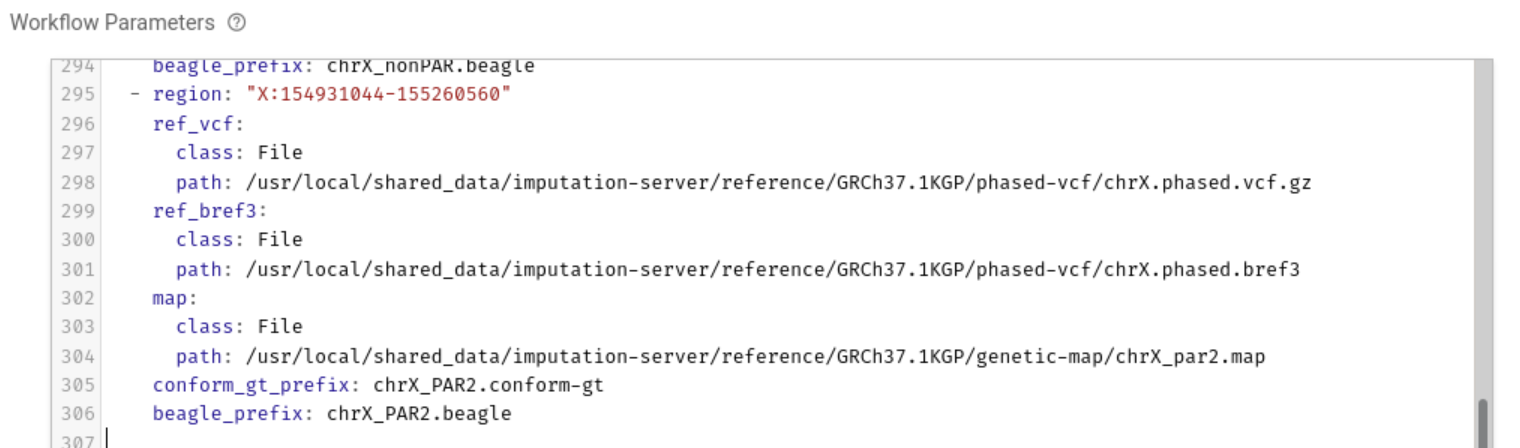
In Workflow Parameters, enter the parameters generated by imputationserver-web-uio.
In this case, delete the {} written from the beginning and enter the generated parameters.
Press the Execute button at the bottom to run the workflow. The status of the job will be Running.
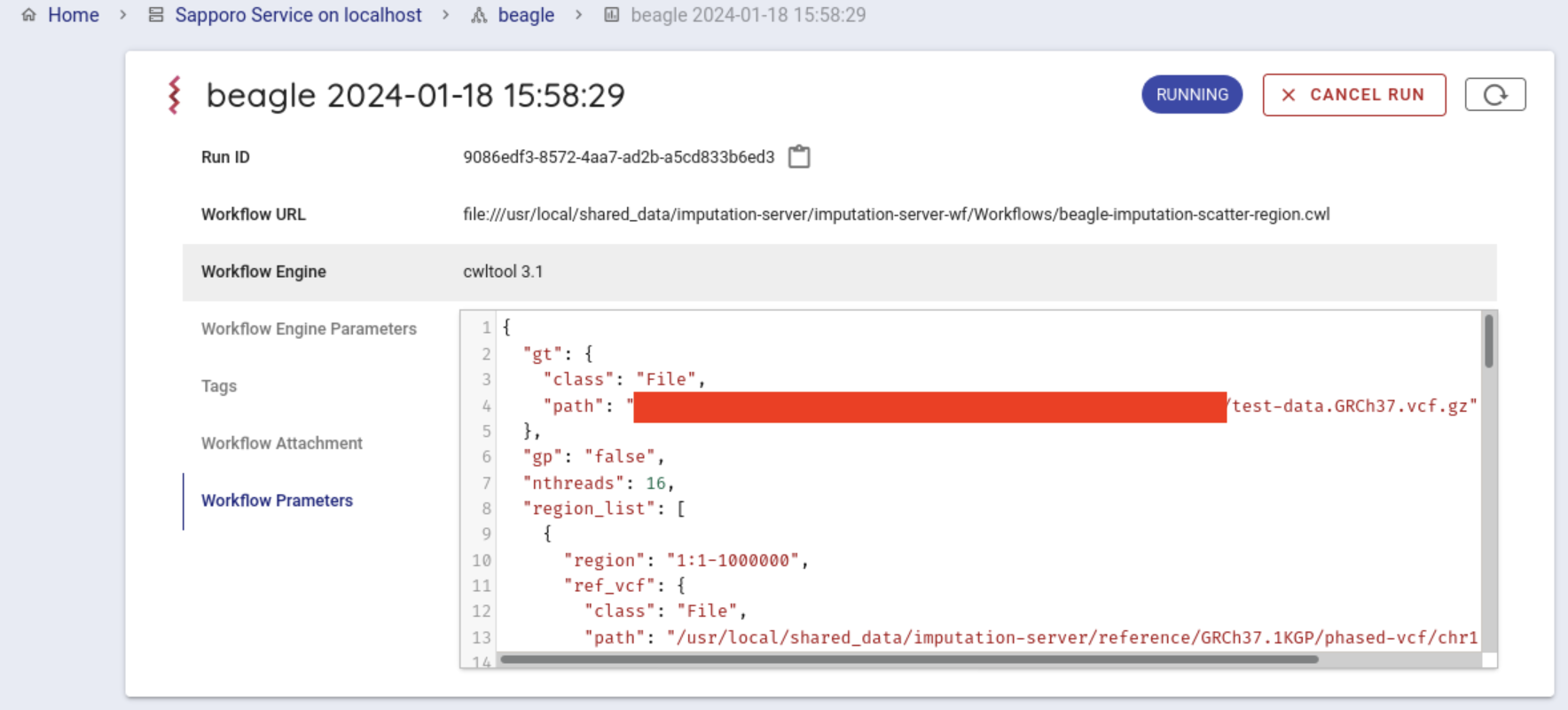
If the workflow is started successfully, the workflow will be run by cwltool.
If successfully completed, COMPLETE.
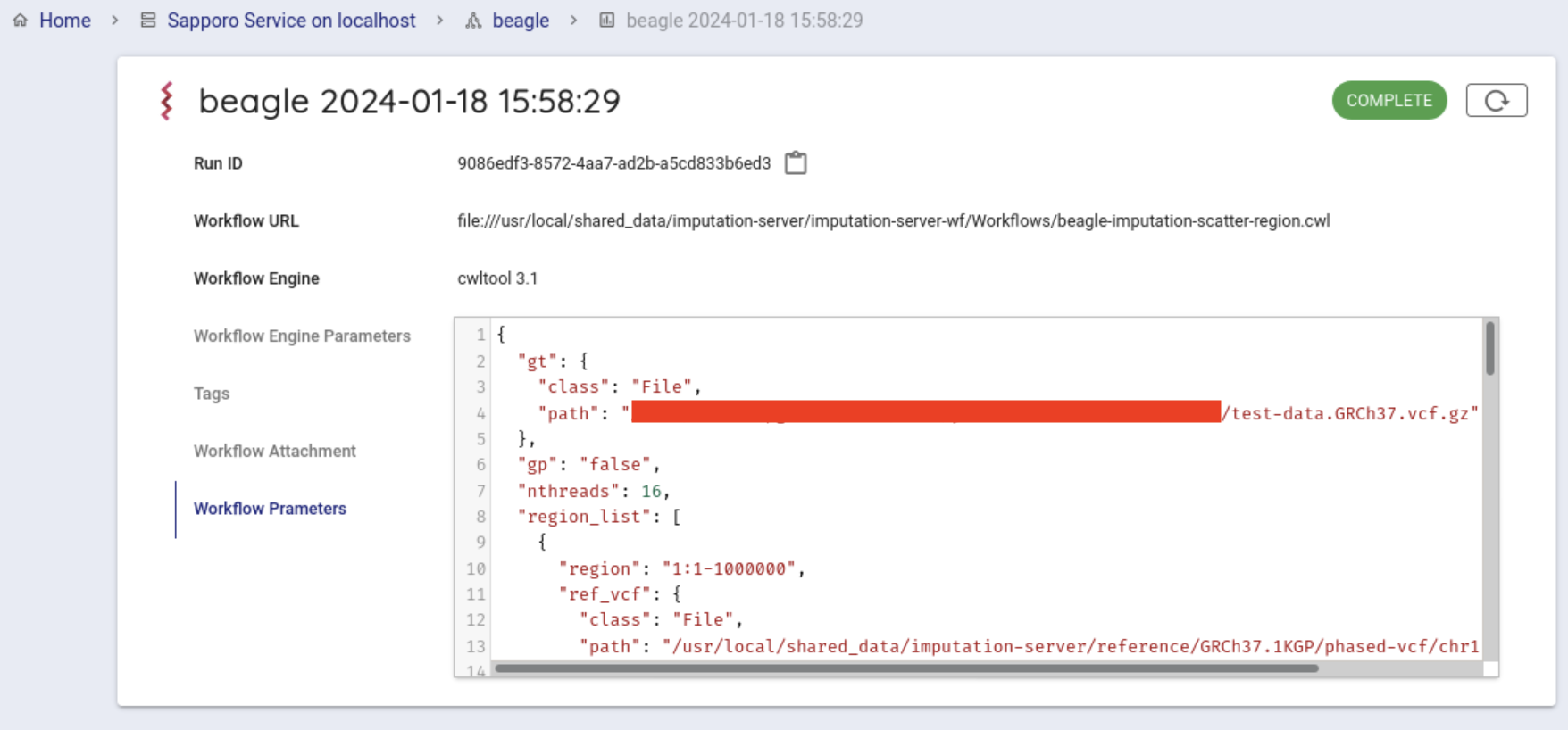
You can get the result file from your browser. Click on Outputs in the Run log to list the result files.
When you click on the file you want to download, a dialogue appears. By default, the file is downloaded under ~/downloads.
Get results
After running the Imputation Workflow, you can get the follows from your web browser.
You can copy the following commands to your computer.
Open a terminal.
When executed, the file will be downloaded to the directory where you are currently executing the command.
scp ([your account name])@gwa.ddbj.nig.ac.jp:~/download/([filename you want to download]) .
(your account name)is the account you use to login to the Personal Genome Analysis environment- For
(file name you want to download), specify the name of the file you want to download.
You can also download the file directly from the results directory of sapporo-service.
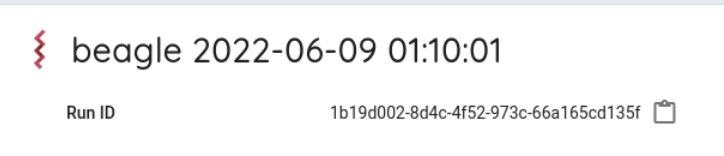
Search Run ID.
The Run ID is displayed on the right of Run ID.
You can copy the Run ID(runid) by clicking on the icon on the right.
All files are in first two characters /runid/outputs/ of the installed directory /sapporo-service/run/runid.
If runid is 1b19d002-8d4c-4f52-973c-66a165cd135f, the first two characters are 1b.
When you copy with the scp command, enter the following.
A directory called outputs will be created in your computer, and the analysis results will be copied from the Personal Genome Analysis division to your computer.
scp -i [your private key file] -r ([your account name])@gwa.ddbj.nig.ac.jp:~/sapporo-install/sapporo-service/run/1b/1b19d002-8d4c-4f52-973c-66a165cd135f/ outputs outputs