Topics before 2022
NBDC-DDBJ Imputation Server (beta)
18 Oct 2022.
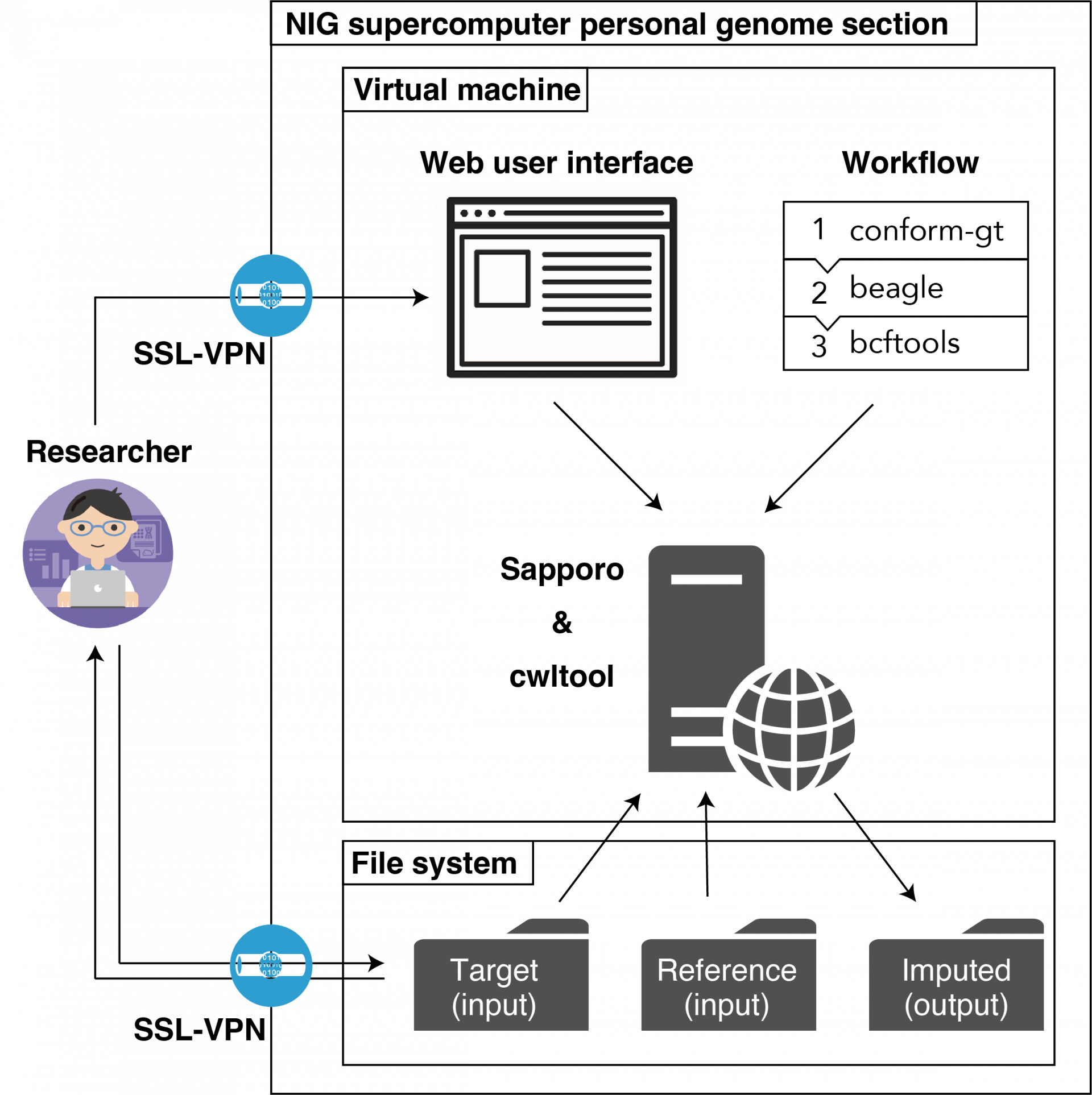
The Imputation Server is a service to support imputation analysis of SNP array data. 🔗Michigan Imputation Server and 🔗TOPMed Project Imputation Server are public. These servers are located outside Japan and genomic data (SNP array data) had to be uploaded to the servers outside Japan for use.
Therefore, 🔗the Department of NBDC Program of the Japan Science and Technology Agency has developed the NBDC-DDBJ Imputation Server system as a Japanese version of the imputation server that is easy for Japanese researchers to use. This system is currently available in the Personal Genome Analysis division of the NIG supercomputer system.
The imputation workflow used in this server was modified and implemented as a web service by the Department of NBDC Program with reference to the information (selection of imputation software and setting of parameters) provided by the National Center for Global Health and Medicine in the following AMED project The NBDC Business Promotion Department has modified and implemented it as a web service using this information as a reference.
Project name: Platform Program for Promotion of Genome Medicine (Research and development research to resolve issues related to international data sharing)
Subject name: "Investigation and practice of ethical and technical issues in genomic medical science research using cloud computing environment"
| The NBDC-DDBJ Imputation Server (beta version) (hereafter referred to as 'this system') is available in the Personal Genome Analysis division of the NIG supercomputer. Researchers(users) can upload their own genome data to the server and execute the imputation analysis workflow via the web user interface. |
PortablePipeline
10 May 2022.
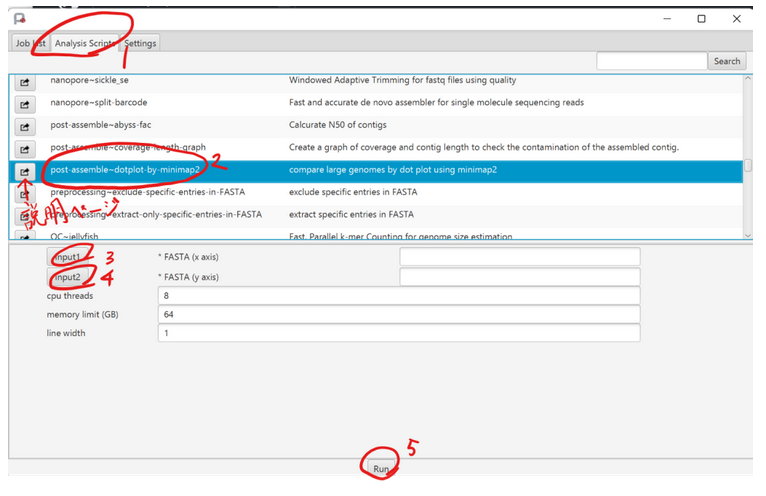
For the NGS analysis pipeline, the tool called Portablepipeline has been developed by Professor Kazutoshi Yoshitake of the Laboratory of Aquatic Molecular Biology and Biotechnology, Aquatic Bioscience, Graduate school of Agricultural and Life Sciences, The Univresity of Tokyo.
For instructions on how to run the tool, refer 🔗 Laboratory of Aquatic Molecular Biology and Biotechnology page.
| "PortablePipeline is software that allows Windows and Mac users to perform NGS analysis in a GUI on a local or remote server or supercomputer. If python3 and docker or singularity are installed as an analysis server, you run it instead of the supercomputer."(Referenced from the Laboratory of Aquatic Molecular Biology and Biotechnology) |
DFAST
2 Oct 2022.
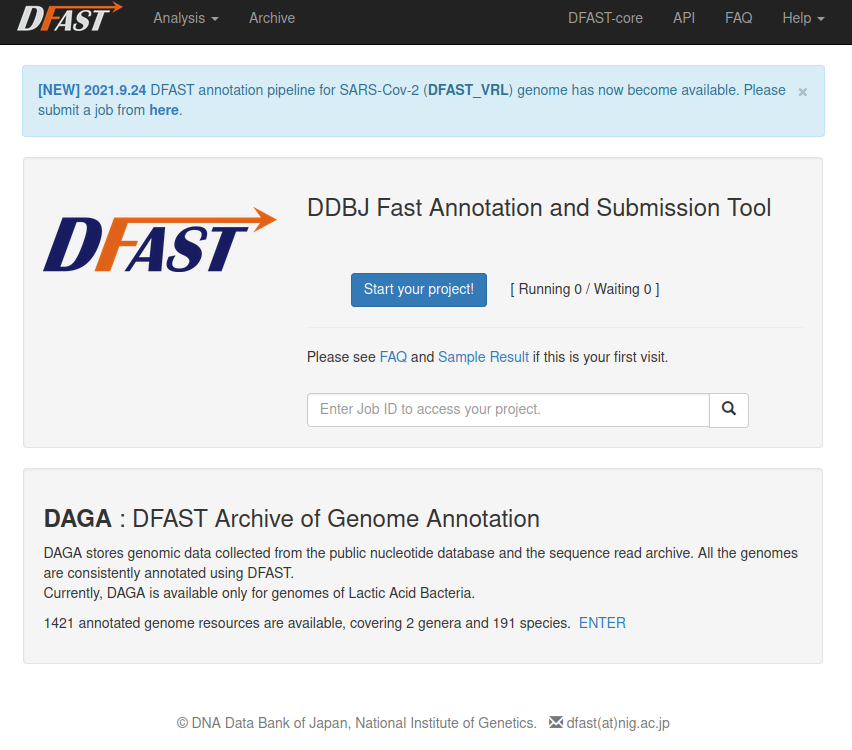
DFAST is an automated annotation tool for prokaryotic genome created by Assistant Professor, Yasuhiro Tanizawa of the Nakamura Group・Genome Informatics Laboratory at the National Institute of Genetics. This tool can also generate files for genome sequence registration to DDBJ. There is a web version available at https://dfast.ddbj.nig.ac.jp that can be used simply by uploading files, and a stand-alone version that can be run by command operation.
| To run the stand-alone version on the NIG supercomputer, there are two ways: get the source code from 🔗https://github.com/nigyta/dfast_core/ and install it (requires Python 3.6 or later and Biopython) or use the singularity container provided by the NIG supercomputer (ref: 🔗https://qiita.com/nigyta/items/e1de21f6ece65d69ec1d). |
Rhelixa RNAseq pipeline
08 Sep 2020.
The RNA-seq analysis pipeline has been provided by 🔗Rhelixa Corporation (CTO: Ryu Nakaki), which has a comprehensive collaboration agreement with the National Institute of Genetics, and implemented on the NIG supercomputer system.
This pipeline maps the Sequence Read Archive of a single sample obtained by the RNA-seq application to a reference genome, aggregates them by gene region and calculates the expression levels of all genes.
| The RNAseq pipeline of Rhelixa is installed on the NIG supercomputer in the form of the Singularity container image. The Singularity container image cannot be used outside the NIG supercomputer. Check to the link below for instructions on how to use it. |