R
This article explains the methods for installing the R environment on the user environment of the National Institute of Genetics supercomputer and using package management. It covers the following topics:
- How to use R included with the OS distribution
- How to build and install R from source code into the user environment
- How to install R using conda
- Managing R analysis environments with renv
- Using the package manager spack
- (Reference) Ways to Speed Up R
How to Use R Included with the OS Distribution
On compute nodes with Ubuntu 22.04 installed, R that comes with Ubuntu 22.04 is pre-installed. You can launch the R environment by entering the R command.
username@at138:~$ R
R version 4.1.2 (2021-11-01) -- "Bird Hippie"
Copyright (C) 2021 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
>
As of February 2024, the installed version of R is 4.1.2 (2021-11-01).
Additionally, the r-cran packages that come with Ubuntu 22.04 are installed.
apt search r-cran
You can refer to the installed package group by entering the above command. Furthermore, after launching R, you can check the libraries available in the R environment by typing:
library()
Other necessary libraries can be individually introduced into the user environment through any of the methods mentioned on this page (such as renv).
How to Install R from a Tarball
This method involves downloading the source code from the R Network, building it, and installing it under the user's home directory. Execute the following to install the R system under $HOME/local/. For the latest source code (tarball) of R, please refer to The Comprehensive R Archive Network.
mkdir -p ~/local/src
cd ~/local/src
# Install R
R_VERSION=4.3.2
R_MAJOR=4
wget https://cran.r-project.org/src/base/R-${R_MAJOR}/R-${R_VERSION}.tar.gz
tar xzvf R-${R_VERSION}.tar.gz
cd R-${R_VERSION}
./configure --prefix=$HOME/local
make
make install
How to Install R with conda
This section describes how to install the R environment using miniconda. For installation of miniconda itself, refer to "How to Use Python" inside Miniconda.
Once miniconda is installed and the conda command is available, execute the following commands.
With conda, it is possible to create and switch between virtual environments for different versions of R.
Check the versions of R available on the channel with the following command:
conda search -c conda-forge r-base
The available versions will be displayed as follows:
yxxxx@at139:~$ conda search -c conda-forge r-base
Loading channels: done
# Name Version Build Channel
r-base 3.3.1 1 pkgs/r
(omission)
r-base 4.3.2 hb8ee39d_1 conda-forge
r-base 4.3.2 hb8ee39d_2 conda-forge
Create an environment for the R version you want to use.
To create an R environment named R-4.3.2, execute the following command. Here, 4.3.2 is just an example; specify the version you wish to use.
xxxx@at139:~$ conda create -n R-4.3.2 -y -c conda-forge r-base=4.3.2
Looking for: ['r-base=4.3.2']
(omission)
conda-forge/linux-64 32.4MB @ 48.6MB/s 0.7s
Transaction
Prefix: /home/xxxx/miniconda3/envs/R-4.3.2
Updating specs:
- r-base=4.3.2
Package Version Build Channel Size
──────────────────────────────────────────────────────────────────────────────────────────
Install:
──────────────────────────────────────────────────────────────────────────────────────────
+ font-ttf-dejavu-sans-mono 2.37 hab24e00_0 conda-forge 397kB
+ font-ttf-inconsolata 3.000 h77eed37_0 conda-forge 97kB
(omission)
+ r-base 4.3.2 hb8ee39d_2 conda-forge 26MB
Summary:
Install: 88 packages
Total download: 184MB
──────────────────────────────────────────────────────────────────────────────────────────
font-ttf-inconsolata 96.5kB @ 1.0MB/s 0.1s
(omission)
libstdcxx-devel_linux-64 13.0MB @ 8.5MB/s 0.4s
sysroot_linux-64 15.3MB @ 9.7MB/s 0.3s
binutils_impl_linux-64 5.4MB @ 3.1MB/s 0.3s
Downloading and Extracting Packages:
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
To activate this environment, use
$ conda activate R-4.3.2
To deactivate an active environment, use
$ conda deactivate
The notation r-base=4.3.2 signifies installing a specific version.
Activate the R environment.
conda activate R-4.3.2
Verify that the prompt now includes (R-4.3.2). Install r-essentials.
conda install -c conda-forge r-essentials
Installation will begin. This may take some time.
Channels:
- conda-forge
- defaults
Platform: linux-64
Collecting package metadata (repodata.json): done
Solving environment: done
## Package Plan ##
environment location: /home/yxxxx/miniconda3/envs/R-4.3.2
added / updated specs:
- r-essentials
The following packages will be downloaded:
package | build
---------------------------|-----------------
anyio-4.2.0 | pyhd8ed1ab_0 99 KB conda-forge
(omission)
zipp-3.17.0 | pyhd8ed1ab_0 19 KB conda-forge
------------------------------------------------------------
Total: 202.4 MB
The following NEW packages will be INSTALLED:
anyio
conda-forge/noarch::anyio-4.2.0-pyhd8ed1ab_0
(omission)
zipp conda-forge/noarch::zipp-3.17.0-pyhd8ed1ab_0
When prompted, enter y.
Proceed ([y]/n)? y
The installation will start.
Downloading and Extracting Packages:
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
(R-4.3.2) xxxx@at137:~$
After the prompt returns, verify that R (4.3.2) launches.
(R-4.3.2) xxxx@at137:~$ R
R version 4.3.2 (2023-10-31) -- "Eye Holes"
Copyright (C) 2023 The R Foundation for Statistical Computing
Platform: x86_64-conda-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
>
To exit the R environment, enter q().
q()
To leave the conda virtual environment, execute conda deactivate.
conda deactivate
You will return to the normal shell environment. If you have entered multiple virtual environments, execute conda deactivate until you exit to the shell.
After installing the R virtual environment once, in subsequent sessions, check the list of previously created virtual environments with
conda env list
and activate the desired virtual environment with conda activate, then launch R.
For subcommands in the conda environment, please refer to the documentation from the developers below.
Once you have installed the R virtual environment, for package management on R, please use renv as described below.
Managing R Analysis Environment with renv
renv is an open-source package for R package management, supported by posit. It was developed to ensure the reproducibility of analysis environments in R.
- renv's GitHub page: https://rstudio.github.io/renv/
- Getting Started with renv: https://rstudio.github.io/renv/articles/renv.html
- renv Function Reference: https://rstudio.github.io/renv/reference/index.html
The workflow for using renv in the R environment is as follows:
- Execute
renv::init(). - Write R code. Install necessary packages using renv.
- During the work process, periodically perform
renv::snapshot()and save it in the lock file. - After finishing the analysis work, execute
renv::snapshot()and save the edited files, renv/, and renv.lock file in an external repository with git commit.
This workflow allows you to save the analysis environment in a repository and reproduce the analysis environment. Below is a diagram summarizing the relationship between renv functions, system library, project library, and lock file.
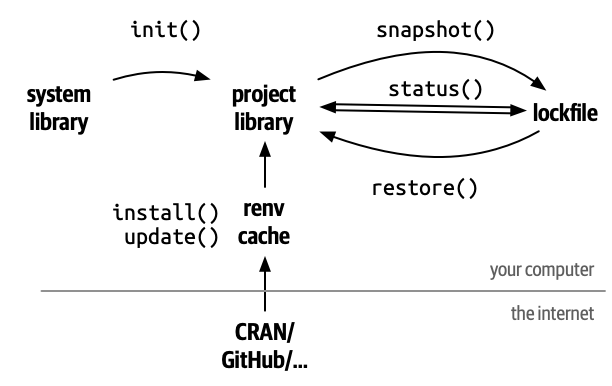
The main renv functions are as follows:
| renv Function | Meaning | Online Manual |
|---|---|---|
| renv::init() | Start package management (execute in a working directory created under the home directory, not directly under the home directory, to avoid errors) | renv::init |
| renv::install() | Install packages | renv::install |
| renv::status() | Check installation status | renv::status |
| renv::snapshot() | Record installation status | renv::snapshot |
| renv::restore() | Reproduce state from renv.lock file | renv::restore |
| renv::history() | View commit history | renv::history |
| renv:: |
revert() | Recover renv.lock using history's commit history | renv::revert | | renv::update() | Update project library | renv::update |
Installing renv
To install the renv package in the R environment, follow these steps:
> install.packages("renv")
Installing package into ‘/home/yxxxx/R/x86_64-pc-linux-gnu-library/4.3’
(as ‘lib’ is unspecified)
trying URL 'https://p3m.dev/cran/__linux__/jammy/latest/src/contrib/renv_1.0.4.tar.gz'
Content type 'binary/octet-stream' length 2102571 bytes (2.0 MB)
==================================================
downloaded 2.0 MB
* installing *binary* package ‘renv’ ...
* DONE (renv)
The downloaded source packages are in
‘/tmp/Rtmp6nfjdw/downloaded_packages’
>
Make the renv package available for use.
> library(renv)
Attaching package: ‘renv’
The following objects are masked from ‘package:stats’:
embed, update
The following objects are masked from ‘package:utils’:
history, upgrade
The following objects are masked from ‘package:base’:
autoload, load, remove
>
Check the installed version of renv.
> print(packageVersion("renv"))
[1] ‘1.0.4’
>
With the above steps, the renv package has been applied.
Example Execution
The following is an execution example on the RStudio Server console. The project is created with the name "renv-test". Executing renv::init() directly under the home directory results in an error, so create a project and then execute init within the project directory.
> renv::init()
The following package(s) will be updated in the lockfile:
# RSPM -----------------------------------------------------------------------
- renv [* -> 1.0.4]
The version of R recorded in the lockfile will be updated:
- R [* -> 4.3.2]
- Lockfile written to "~/renv-test/renv.lock".
Restarting R session...
- Project '~/renv-test' loaded. [renv 1.0.4]
Try installing a package.
> install.packages("tidyr")
# Downloading packages -------------------------------------------------------
- Downloading tidyr from CRAN ... OK [1.1 Mb in 1.3s]
- Downloading cli from CRAN ... OK [1.2 Mb in 0.96s]
- Downloading dplyr from CRAN ... OK [1.4 Mb in 1.0s]
- Downloading generics from CRAN ... OK [74.5 Kb in 0.93s]
- Downloading glue from CRAN ... OK [146.4 Kb in 0.96s]
- Downloading lifecycle from CRAN ... OK [120.5 Kb in 0.93s]
- Downloading rlang from CRAN ... OK [1.5 Mb in 1.1s]
- Downloading magrittr from CRAN ... OK [215.9 Kb in 0.93s]
- Downloading pillar from CRAN ... OK [631.4 Kb in 0.97s]
- Downloading fansi from CRAN ... OK [300.6 Kb in 1.1s]
- Downloading utf8 from CRAN ... OK [143.4 Kb in 0.94s]
- Downloading vctrs from CRAN ... OK [1.2 Mb in 0.94s]
- Downloading R6 from CRAN ... OK [80.6 Kb in 0.97s]
- Downloading tibble from CRAN ... OK [656.1 Kb in 0.95s]
- Downloading pkgconfig from CRAN ... OK [17.2 Kb in 1.0s]
- Downloading tidyselect from CRAN ... OK [214.8 Kb in 0.93s]
- Downloading withr from CRAN ... OK [232.4 Kb in 0.93s]
- Downloading purrr from CRAN ... OK [477.1 Kb in 0.97s]
- Downloading stringr from CRAN ... OK [298.1 Kb in 0.97s]
- Downloading stringi from CRAN ... OK [3.1 Mb in 0.96s]
- Downloading cpp11 from CRAN ... OK [273.7 Kb in 0.94s]
Successfully downloaded 21 packages in 26 seconds.
The following package(s) will be installed:
- cli [3.6.2]
- cpp11 [0.4.7]
- dplyr [1.1.4]
- fansi [1.0.6]
- generics
[0.1.3]
- glue [1.7.0]
- lifecycle [1.0.4]
- magrittr [2.0.3]
- pillar [1.9.0]
- pkgconfig [2.0.3]
- purrr [1.0.2]
- R6 [2.5.1]
- rlang [1.1.3]
- stringi [1.8.3]
- stringr [1.5.1]
- tibble [3.2.1]
- tidyr [1.3.1]
- tidyselect [1.2.0]
- utf8 [1.2.4]
- vctrs [0.6.5]
- withr [3.0.0]
These packages will be installed into "~/renv-test/renv/library/R-4.3/x86_64-pc-linux-gnu".
Do you want to proceed? [Y/n]: y
Enter y.
# Installing packages --------------------------------------------------------
- Installing cli ... OK [installed binary and cached in 1.3s]
- Installing generics ... OK [installed binary and cached in 0.56s]
- Installing glue ... OK [installed binary and cached in 0.89s]
- Installing rlang ... OK [installed binary and cached in 1.4s]
- Installing lifecycle ... OK [installed binary and cached in 1.2s]
- Installing magrittr ... OK [installed binary and cached in 0.97s]
- Installing fansi ... OK [installed binary and cached in 0.92s]
- Installing utf8 ... OK [installed binary and cached in 0.7s]
- Installing vctrs ... OK [installed binary and cached in 1.4s]
- Installing pillar ... OK [installed binary and cached in 1.5s]
- Installing R6 ... OK [installed binary and cached in 0.7s]
- Installing pkgconfig ... OK [installed binary and cached in 0.64s]
- Installing tibble ... OK [installed binary and cached in 1.7s]
- Installing withr ... OK [installed binary and cached in 0.87s]
- Installing tidyselect ... OK [installed binary and cached in 0.95s]
- Installing dplyr ... OK [installed binary and cached in 1.9s]
- Installing purrr ... OK [installed binary and cached in 0.99s]
- Installing stringi ... OK [installed binary and cached in 0.96s]
- Installing stringr ... OK [installed binary and cached in 1.4s]
- Installing cpp11 ... OK [installed binary and cached in 1.2s]
- Installing tidyr ... OK [installed binary and cached in 1.6s]
Successfully installed 21 packages in 25 seconds.
After installing packages, perform your R coding tasks. Once completed, run renv::snapshot().
> renv::snapshot()
The following package(s) will be updated in the lockfile:
# CRAN -----------------------------------------------------------------------
- cli [* -> 3.6.2]
- cpp11 [* -> 0.4.7]
- dplyr [* -> 1.1.4]
- fansi [* -> 1.0.6]
- generics [* -> 0.1.3]
- glue [* -> 1.7.0]
- lifecycle [* -> 1.0.4]
- magrittr [* -> 2.0.3]
- pillar [* -> 1.9.0]
- pkgconfig [* -> 2.0.3]
- purrr [* -> 1.0.2]
- R6 [* -> 2.5.1]
- rlang [* -> 1.1.3]
- stringi [* -> 1.8.3]
- stringr [* -> 1.5.1]
- tibble [* -> 3.2.1]
- tidyr [* -> 1.3.1]
- tidyselect [* -> 1.2.0]
- utf8 [* -> 1.2.4]
- vctrs [* -> 0.6.5]
- withr [* -> 3.0.0]
Do you want to proceed? [Y/n]:
Enter y.
- Lockfile written to "~/renv-test/renv.lock".
>
The lockfile is written. Managing this renv.lock file in a git repository, for instance, allows you to restore the library environment
on another host or project folder using renv::restore().
How to Use the Package Manager Spack
Spack is one of the package managers that can be used with user privileges only.
Basic Procedure
Please install the Spack package manager according to the instructions in Installing Spack.
Executing the following commands will make the R environment available.
spack install r
spack load r
Detailed Installation Instructions
Displaying Available Packages
The package name for the R environment is simply r, so some ingenuity is required to search for available R-related packages. Executing spack list | grep ^r | less will display R-related packages.
$ spack list | grep ^r | less
r
r-a4
r-a4base
r-a4classif
r-a4core
r-a4preproc
r-a4reporting
r-abadata
r-abaenrichment
r-abind
... (omitted)
Displaying Available Versions
Since the package name for the R environment itself is r, executing spack info r will display available versions of R.
$ spack info r
AutotoolsPackage: r
Description:
R is 'GNU S', a freely available language and environment for
statistical computing and graphics which provides a wide variety of
statistical and graphical techniques: linear and nonlinear modelling,
statistical tests, time series analysis, classification, clustering,
etc. Please consult the R project homepage for further information.
Homepage: https://www.r-project.org
Preferred version:
4.1.3 https://cloud.r-project.org/src/base/R-4/R-4.1.3.tar.gz
Safe versions:
4.1.3 https://cloud.r-project.org/src/base/R-4/R-4.1.3.tar.gz
4.1.2 https://cloud.r-project.org/src/base/R-4/R-4.1.2.tar.gz
4.1.1 https://cloud.r-project.org/src/base/R-4/R-4.1.1.tar.gz
4.1.0 https://cloud.r-project.org/src/base/R-4/R-4.1.0.tar.gz
4.0.5 https://cloud.r-project.org/src/base/R-4/R-4.0.5.tar.gz
4.0.4 https://cloud.r-project.org/src/base/R-4/R-4.0.4.tar.gz
4.0.3 https://cloud.r-project.org/src/base/R-4/R-4.0.3.tar.gz
4.0.2 https://cloud.r-project.org/src/base/R-4/R-4.0.2.tar.gz
4.0.1 https://cloud.r-project.org/src/base/R-4/R-4.0.1.tar.gz
4.0.0 https://cloud.r-project.org/src/base/R-4/R-4.0.0.tar.gz
3.6.3 https://cloud.r-project.org/src/base/R-3/R-3.6.3.tar.gz
3.6.2 https://cloud.r-project.org/src/base/R-3/R-3.6.2.tar.gz
3.6.1 https://cloud.r-project.org/src/base/R-3/R-3.6.1.tar.gz
3.6.0 https://cloud.r-project.org/src/base/R-3/R-3.6.0.tar.gz
3.5.3 https://cloud.r-project.org/src/base/R-3/R-3.5.3.tar.gz
3.5.2 https://cloud.r-project.org/src/base/R-3/R-3.5.2.tar.gz
3.5.1 https://cloud.r-project.org/src/base/R-3/R-3.5.1.tar.gz
3.5.0 https://cloud.r-project.org/src/base/R-3/R-3.5.0.tar.gz
3.4.4 https://cloud.r-project.org/src/base/R-3/R-3.4.4.tar.gz
3.4.3 https://cloud.r-project.org/src/base/R-3/R-3.4.3.tar.gz
3.4.2 https://cloud.r-project.org/src/base/R-3/R-3.4.2.tar.gz
3.4.1 https://cloud.r-project.org/src/base/R-3/R-3.4.1.tar.gz
3.4.0 https://cloud.r-project.org/src/base/R-3/R-3.4.0.tar.gz
3.3.3 https://cloud.r-project.org/src/base/R-3/R-3.3.3.tar.gz
3.3.2 https://cloud.r-project.org/src/base/R-3/R-3.3.2.tar.gz
3.3.1 https://cloud.r-project.org/src/base/R-3/R-3.3.1.tar.gz
3.3.0 https://cloud.r-project.org/src/base/R-3/R-3.3.0.tar.gz
3.2.5 https://cloud.r-project.org/src/base/R-3/R-3.2.5.tar.gz
3.2.3 https://cloud.r-project.org/src/base/R-3/R-3.2.3.tar.gz
3.2.2 https://cloud.r-project.org/src/base/R-3/R-3.2.2.tar.gz
3.2.1 https://cloud.r-project.org/src/base/R-3/R-3.2.1.tar.gz
3.2.0 https://cloud.r-project.org/src/base/R-3/R-3.2.0.tar.gz
3.1.3 https://cloud.r-project.org/src/base/R-3/R-3.1.3.tar.gz
3.1.2 https://cloud.r-project.org/src/base/R-3/R-3.1.2.tar.gz
Deprecated versions:
None
Variants:
====================== ==== ============== ==================================================
X [off] -- on, off Enable X11 support (TCLTK, PNG, JPEG, TIFF, CAIRO)
external-lapack [off] -- on, off Links to externally installed BLAS/LAPACK
memory_profiling [off] -- on, off Enable memory profiling
rmath [off] -- on, off Build standalone Rmath library
Build Dependencies:
blas cairo gnuconfig icu4c jpeg libpng libx11 libxt pango pcre2 tk zlib
bzip2 curl harfbuzz java lapack libtiff libxmu ncurses pcre readline xz
Link Dependencies:
blas cairo harfbuzz java lapack libtiff libxmu ncurses pcre readline xz
bzip2 curl icu4c jpeg libpng libx11 libxt pango pcre2 tk zlib
Run Dependencies:
None
To install a specific version, execute the command as follows:
spack install r@4.0.5
spack load r@4.0.5
Switching R Versions
Check the currently installed versions.
$ spack find r
==> 2 installed packages
-- linux-centos7-zen2 / intel@2021.4.0 --------------------------
r@4.0.5 r@4.1.3
Loading the desired version with spack load will switch to that version.
$ spack load r@4.0.5
$ R
R version 4.0.5 (2021-03-31) -- "Shake and Throw"
Copyright (C) 2021 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> q()
Save workspace image? [y/n/c]: n
$ spack load r@4.1.3
$ R
R version 4.1.3 (2022-03-10) -- "One Push-Up"
Copyright (C) 2022 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> q()
$
Adjusting Compilation Options
By looking at the Variants section in the spack info r command, you can see that by default, X window related libraries are not linked, and graphical output of graphs is not possible in this state.
Compilation options can be adjusted during spack install as follows (note that compilation time will significantly increase).
spack install r@4.0.5 X=True
Uninstalling
The following command can be used to uninstall packages.
spack uninstall r
For uninstalling Spack itself, please refer to Installing Spack.
(Reference) Ways to Speed Up R
At the Genomic Research Supercomputer, the Intel oneAPI Base & HPC Toolkit Multi-Node has been introduced, and the following are available:
- Intel oneAPI DPC++/C++ Compiler
- Intel MKL (Math Kernel Library)
It is possible to build and speed up R source code using these tools. This section describes how to do that.
Building R with the GNU Compiler and Linking with MKL as the BLAS Library
This section describes how to link R with the Intel MKL as the BLAS library using the gcc and gfortran compilers. Although methods for compiling the source code with the Intel Compiler will be discussed later, using MKL can already lead to performance improvements. This item is based on the following document:
Set the environment variables for configure. This follows the documentation for building with MKL's multi-threaded support. If you want single-threaded support, please adjust accordingly. Here, we're downloading and unpacking the source code package for R-4.3.2 from CRAN.
yxxxx@at138:~/R2/R-4.3.2$ export MKL="-L/lustre7/software/intel_ubuntu/oneapi/mkl/latest/lib/intel64 -lmkl_gf_lp64 -lmkl_core -lmkl_gnu_thread -dl -fopenmp"
yxxxx@at138:~/R2/R-4.3.2$ export MKL_INTERFACE_LAYER=GNU,LP64
yxxxx@at138:~/R2/R-4.3.2$ export MKL_THREADING_LAYER=GNU
Use the above variables for configure. If there are errors related to libcurl, zlib, or iconv, it is suspected that the build is not looking at the OS-provided tools but those inside miniconda, so exit the conda environment and ensure that the miniconda environment is not referenced during the build.
yxxxx@at138:~/R2/R-4.3.2$ ./configure --with-blas="$MKL" --with-lapack --prefix=/home/yxxxx/R-4.3.2MKL
checking build system type
... x86_64-pc-linux-gnu
checking host system type... x86_64-pc-linux-gnu
loading site script './config.site'
loading build-specific script './config.site'
checking for pwd... /usr/bin/pwd
checking whether builddir is srcdir... yes
checking whether ln -s works... yes
checking for ar... ar
checking for a BSD-compatible install... /usr/bin/install -c
(omitted for brevity)
Make sure to check the environment for BLAS during configure as follows.
checking for dgemm_ in -L/lustre7/software/intel_ubuntu/oneapi/mkl/latest/lib/intel64 -lmkl_gf_lp64 -lmkl_core -lmkl_gnu_thread -dl -fopenmp... yes
checking whether double complex BLAS can be used... yes
checking whether the BLAS is complete... yes
(omitted for brevity)
R is now configured for x86_64-pc-linux-gnu
Source directory: .
Installation directory: /home/yxxxx/R-4.3.2MKL
C compiler: gcc -g -O2
Fortran fixed-form compiler: gfortran -g -O2
Default C++ compiler: g++ -std=gnu++17 -g -O2
C++11 compiler: g++ -std=gnu++11 -g -O2
C++14 compiler: g++ -std=gnu++14 -g -O2
C++17 compiler: g++ -std=gnu++17 -g -O2
C++20 compiler: g++ -std=gnu++20 -g -O2
C++23 compiler: g++ -std=gnu++23 -g -O2
Fortran free-form compiler: gfortran -g -O2
Obj-C compiler:
Interfaces supported: X11, tcltk
External libraries: pcre2, readline, BLAS(MKL), LAPACK(in blas), curl
Additional capabilities: PNG, JPEG, TIFF, NLS, cairo, ICU
Options enabled: R profiling
Capabilities skipped:
Options not enabled: shared BLAS, memory profiling
Recommended packages: yes
Please note that BLAS is using MKL.
Then proceed with make, ensuring there are no interruptions due to errors.
yxxxx@at138:~/R2/R-4.3.2$ make
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/m4'
(omitted for brevity)
configuring Java ...
Java interpreter : /usr/bin/java
Java version : 11.0.20.1
Java home path : /usr/lib/jvm/java-11-openjdk-amd64
Java compiler : /usr/bin/javac
Java headers gen.:
Java archive tool: /usr/bin/jar
trying to compile and link a JNI program
detected JNI cpp flags : -I$(JAVA_HOME)/include -I$(JAVA_HOME)/include/linux
detected JNI linker flags : -L$(JAVA_HOME)/lib/server -ljvm
using C compiler: ‘gcc (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0’
make[2]: Entering directory '/tmp/Rjavareconf.qqHvXL'
gcc -I"/home/yxxxx/R2/R-4.3.2/include" -DNDEBUG -I/usr/lib/jvm/java-11-openjdk-amd64/include -I/usr/lib/jvm/java-11-openjdk-amd64/include/linux -I/usr/local/include -fpic -g -O2 -c conftest.c -o conftest.o
gcc -shared -L/usr/local/lib -o conftest.so conftest.o -L/usr/lib/jvm/java-11-openjdk-amd64/lib/server -ljvm
make[2]: Leaving directory '/tmp/Rjavareconf.qqHvXL'
JAVA_HOME : /usr/lib/jvm/java-11-openjdk-amd64
Java library path: $(JAVA_HOME)/lib/server
JNI cpp flags : -I$(JAVA_HOME)/include -I$(JAVA_HOME)/include/linux
JNI linker flags : -L$(JAVA_HOME)/lib/server -ljvm
Updating Java configuration in /home/yxxxx/R2/R-4.3.2
Done.
Proceed to make install.
yxxxx@at138:~/R2/R-4.3.2$ make install
mkdir -p -- /home/yxxxx/R
-4.3.2MKL/lib/R
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/m4'
make[1]: Nothing to be done for 'install'.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/m4'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/tools'
(omitted for brevity)
gcc -I. -I../../src/include -I../../src/include -I/usr/local/include -DHAVE_CONFIG_H -g -O2 -L/usr/local/lib -DR_HOME='"/home/yxxxx/R-4.3.2MKL/lib/R"' \
-o Rscript ./Rscript.c
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/unix'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/main'
mkdir -p -- /home/yxxxx/R-4.3.2MKL/lib/R/bin/exec
make[3]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/main'
make[3]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/main'
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/main'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules'
make[3]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules/internet'
mkdir -p -- /home/yxxxx/R-4.3.2MKL/lib/R/modules
make[3]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules/internet'
make[3]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules/lapack'
make[3]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules/lapack'
make[3]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules/X11'
make[3]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules/X11'
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/library'
mkdir -p -- /home/yxxxx/R-4.3.2MKL/lib/R/library
installing packages ...
building HTML index ...
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/library'
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/tests'
make[1]: Nothing to be done for 'install'.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/tests'
Verify that R has been installed in the target directory.
yxxxx@at138:~/R-4.3.2MKL/bin$ pwd
/home/yxxxx/R-4.3.2MKL/bin
yxxxx@at138:~/R-4.3.2MKL/bin$ ls -l R
-rwxr-xr-x 1 yxxxx co-xxx 9162 Mar 16 10:07 R
yxxxx@at138:~/R-4.3.2MKL/bin$ ./R
R version 4.3.2 (2023-10-31) -- "Eye Holes"
Copyright (C) 2023 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
The R built this time operates in multi-thread mode, so set the number of cores to
use with the environment variable OMP_NUM_THREADS or MKL_NUM_THREADS before running R. Using too many cores may lead to excessive CPU resource usage and parallel operation overhead, potentially resulting in worse performance or even deadlocks. Please specify a reasonable number of cores based on the problem size and program operation.
Performance Comparison Between Using and Not Using MKL (Reference)
Here we present some simple benchmark results.
Since these benchmarks were run on a node shared by multiple users with multiple programs running, the results are for reference only. The R version using MKL had MKL_NUM_THREADS set to 5. The version of R used was 4.3.2.
Without Using MKL (Not Using External BLAS)
yxxxx@at139:~/R-4.3.2MKL/bin$ cat R-benchmark-25.R |~/R-plane/bin/R --slave
Loading required package: Matrix
Loading required package: SuppDists
Warning message:
In library(package, lib.loc = lib.loc, character.only = TRUE, logical.return = TRUE, :
there is no package called ‘SuppDists’
Warning messages:
1: In remove("a", "b") : object 'a' not found
2: In remove("a", "b") : object 'b' not found
R Benchmark 2.5
===============
Number of times each test is run__________________________: 3
I. Matrix calculation
---------------------
Creation, transposition, deformation of a 2500x2500 matrix (sec): 0.497
2400x2400 normally distributed random matrix ^1000____ (sec): 0.338
Sorting of 7,000,000 random values__________________ (sec): 0.783333333333333
2800x2800 cross-product matrix (b = a' * a)_________ (sec): 10.3113333333333
Linear regression over a 3000x3000 matrix (c = a \ b')___ (sec): 3.76533333333333
--------------------------------------------
Trimmed geometric mean (2 extremes eliminated): 1.13597509074229
II. Matrix functions
--------------------
FFT over 2,400,000 random values____________________ (sec): 0.186333333333332
Eigenvalues of a 640x640 random matrix______________ (sec): 0.592666666666664
Determinant of a 2500x2500 random matrix____________ (sec): 0.184666666666667
Cholesky decomposition of a 3000x3000 matrix________ (sec): 0.0586666666666673
Inverse of a 1600x1600 random matrix________________ (sec): 1.672
--------------------------------------------
Trimmed geometric mean (2 extremes eliminated): 0.273209956216133
III. Programming
------------------
3,500,000 Fibonacci numbers calculation (vector calc)(sec): 0.175333333333332
Creation of a 3000x3000 Hilbert matrix (matrix calc) (sec): 0.184666666666667
Greatest common divisors of 400,000 pairs (recursion)__ (sec): 0.147666666666666
Creation of a 500x500 Toeplitz matrix (loops)_______ (sec): 0.042666666666662
Escoufier's method on a 45x45 matrix (mixed)________ (sec): 0.307999999999993
--------------------------------------------
Trimmed geometric mean (2 extremes eliminated): 0.168465829089985
Total time for all 15 tests_________________________ (sec): 19.2476666666667
Overall mean (sum of I, II, and III trimmed means/3)_ (sec): 0.373931786643322
--- End of test ---
With MKL
yxxxx@at139:~/R-4.3.2MKL/bin$ cat R-benchmark-25.R |./R --slave
Loading required package: Matrix
Loading required package: SuppDists
Warning message:
In library(package, lib.loc = lib.loc, character.only = TRUE, logical.return = TRUE, :
there is no package called ‘SuppDists’
Warning messages:
1: In remove("a", "b") : object 'a' not found
2: In remove("a", "b") : object 'b' not found
R Benchmark 2.5
===============
Number of times each test is run__________________________: 3
I. Matrix calculation
---------------------
Creation, transposition, deformation of a 2500x2500 matrix (sec): 0.511333333333334
2400x2400 normally distributed random matrix ^1000____ (sec): 0.337666666666667
Sorting of 7,000,000 random values__________________ (sec): 0.799666666666667
2800x2800 cross-product matrix (b = a' * a)_________ (sec): 0.148666666666666
Linear regression over a 3000x3000 matrix (c = a \ b')___ (sec): 0.107999999
999999
--------------------------------------------
Trimmed geometric mean (2 extremes eliminated): 0.294986381459619
II. Matrix functions
--------------------
FFT over 2,400,000 random values____________________ (sec): 0.186
Eigenvalues of a 640x640 random matrix______________ (sec): 0.223666666666666
Determinant of a 2500x2500 random matrix____________ (sec): 0.0846666666666659
Cholesky decomposition of a 3000x3000 matrix________ (sec): 0.074333333333333
Inverse of a 1600x1600 random matrix________________ (sec): 0.120000000000001
--------------------------------------------
Trimmed geometric mean (2 extremes eliminated): 0.123633325394564
III. Programming
------------------
3,500,000 Fibonacci numbers calculation (vector calc)(sec): 0.173999999999999
Creation of a 3000x3000 Hilbert matrix (matrix calc) (sec): 0.187000000000001
Greatest common divisors of 400,000 pairs (recursion)__ (sec): 0.146000000000001
Creation of a 500x500 Toeplitz matrix (loops)_______ (sec): 0.0439999999999981
Escoufier's method on a 45x45 matrix (mixed)________ (sec): 0.235000000000003
--------------------------------------------
Trimmed geometric mean (2 extremes eliminated): 0.168105234521273
Total time for all 15 tests_________________________ (sec): 3.38
Overall mean (sum of I, II, and III trimmed means/3)_ (sec): 0.18302324510412
--- End of test ---
It can be seen that using MKL results in approximately a two-fold speed increase.
(Reference) How to Compile R with Intel Compiler and Link MKL for Building R
This document describes how to compile R's source code with optimization level 3 using the Intel compiler and link it with MKL, based on the following reference:
As mentioned in the document, it has been confirmed on the Genomic Research Supercomputer that an error occurs during operation checks if -lmkl_sequential is not specified with the latest Intel compiler, and this has not been circumvented. This option indicates the use of the MKL library that does not perform thread parallel operations, hence, currently, it is not possible to benefit from parallel operations. Therefore, the following explains the procedure when the build passes by specifying -lmkl_sequential, for reference only.
First, set the following environment variables for preparation:
CXX=icpx
FC=ifx
CC=icx
CFLAGS=-O3 -fp-model precise -Wall -Wstrict-prototypes
CXXFLAGS=-O3 -fp-model precise -Wall
FFLAGS=-O3 -fp-model precise -warn all,noexternals
LDFLAGS=-L/lustre7/software/intel_ubuntu/oneapi/compiler/2024.0/lib
C17FLAGS=-O3 -fp-model precise -Wall -Wno-strict-prototypes
MKL=-L/lustre7/software/intel_ubuntu/oneapi/mkl/2023.2.0/lib/intel64 -lmkl_intel_lp64 -lmkl_core -lmkl_sequential
FCFLAGS=-free -O3 -fp-model precise -warn all,noexternals
Run configure as follows:
yxxxx@at138:~/R2/R-4.3.2$ ./configure --with-blas="$MKL" --with-lapack --prefix=/home/yxxxx/R-Intel
checking build system type... x86_64-pc-linux-gnu
checking host system type... x86_64-pc-linux-gnu
loading site script './config.site'
loading build-specific script './config.site'
checking for pwd... /usr/bin/pwd
checking whether builddir is srcdir... yes
checking whether ln -s works... yes
checking for ar... ar
(省略)
checking for dgemm_ in -L/lustre7/software/intel_ubuntu/oneapi/mkl/2023.2.0/lib/intel64 -lmkl_intel_lp64 -lmkl_core -lmkl_sequential... yes
checking whether double complex BLAS can be used... yes
checking whether the BLAS is complete... yes
(省略)
R is now configured for x86_64-pc-linux-gnu
Source directory: .
Installation directory: /home/yxxxx/R-Intel
C compiler: icx -O3 -fp-model precise -Wall -Wstrict-prototypes
Fortran fixed-form compiler: ifx -O3 -fp-model precise -warn all,noexternals
Default C++ compiler: icpx -std=gnu++17 -O3 -fp-model precise -Wall
C++11 compiler: icpx -std=gnu++11 -O3 -fp-model precise -Wall
C++14 compiler: icpx -std=gnu++14 -O3 -fp-model precise -Wall
C++17 compiler: icpx -std=gnu++17 -O3 -fp-model precise -Wall
C++20 compiler: icpx -std=gnu++20 -O3 -fp-model precise -Wall
C++23 compiler: icpx -std=gnu++23 -O3 -fp-model precise -Wall
Fortran free-form compiler: ifx -free -O3 -fp-model precise -warn all,noexternals
Obj-C compiler:
Interfaces supported: X11, tcltk
External libraries: pcre2, readline, BLAS(MKL), LAPACK(in blas), curl
Additional capabilities: PNG, JPEG, TIFF, NLS, cairo, ICU
Options enabled: R profiling
Capabilities skipped:
Options not enabled: shared BLAS, memory profiling
Recommended packages: yes
Ensure that the compilers used are not GNU compilers but icpx and ifx, and that BLAS is linked with MKL.
Proceed with make:
yxxxx@at138:~/R2/R-4.3.2$ make
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/m4'
make[1]: Nothing to be done for 'R'.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/m4'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/tools'
make[1]: Nothing to be done for 'R'.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/tools'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/doc'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/doc/html'
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/doc/html'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/doc/manual'
make[2]: Nothing to be done for 'R'.
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/doc/manual'
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/doc'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/etc'
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/etc'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/share'
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/share'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/scripts'
creating src/scripts/R.fe
make[3]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/scripts'
make[3]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/scripts'
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/scripts'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/include'
config.status: creating src/include/config.h
config.status: src/include/config.h is unchanged
make[3]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/include/R_ext'
make[3]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/include/R_ext'
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/include'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/extra'
make[3]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/extra/tre'
make[4]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/extra/tre'
making regcomp.d from regcomp.c
making regerror.d from regerror.c
making regexec.d from regexec.c
making tre-ast.d from tre-ast.c
making tre-compile.d from tre-compile.c
making tre-match-approx.d from tre-match-approx.c
making tre-match-backtrack.d from tre-match-backtrack.c
making tre-match-parallel.d from tre-match-parallel.c
making tre-mem.d from tre-mem.c
making tre-parse.d from tre-parse.c
making tre-stack.d from tre-stack.c
make[4]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/extra/tre'
make[4]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/extra/tre'
icx -I. -I. -I../../../src/include -I../../../src/include -I/usr/local/include -DHAVE_CONFIG_H -fiopenmp -O3 -fp-model precise -Wall -Wstrict-prototypes -c regcomp.c -o regcomp.o
icx -I. -I. -I../../../src/include -I../../../src/include -I/usr/local/include -DHAVE_CONFIG_H -fiopenmp -O3 -fp-model precise -Wall -Wstrict-prototypes -c regerror.c -o regerror.o
icx -I. -I. -I../../../src/include -I../../../src/include -I/usr/local/include -DHAVE_CONFIG_H -fiopenmp -O3 -fp-model precise -Wall -Wstrict-prototypes -c regexec.c -o regexec.o
icx -I. -I. -I../../../src/include -I../../../src/include -I/usr/local/include -DHAVE_CONFIG_H -fiopenmp -O3 -fp-model precise -Wall -Wstrict-prototypes -c tre-ast.c -o tre-ast.o
icx -I. -I. -I../../../src/include -I../../../src/include -I/usr/local/include -DHAVE_CONFIG_H -fiopenmp -O3 -fp-model precise -Wall -Wstrict-prototypes -c tre-compile.c -o tre-compile.o
icx -I. -I. -I../../../src/include -I../../../src/include -I/usr/local/include -DHAVE_CONFIG_H -fiopenmp -O3 -fp-model precise -Wall -Wstrict-prototypes -c tre-match-approx.c -o tre-match-approx.o
(省略)
trying to compile and link a JNI program
detected JNI cpp flags : -I$(JAVA_HOME)/include -I$(JAVA_HOME)/include/linux
detected JNI linker flags : -L$(JAVA_HOME)/lib/server -ljvm
using C compiler: ‘Intel(R) oneAPI DPC++/C++ Compiler 2024.0.0 (2024.0.0.20231017)’
make[2]: Entering directory '/tmp/Rjavareconf.5WK0Ji'
icx -I"/home/yxxxx/R2/R-4.3.2/include" -DNDEBUG -I/usr/lib/jvm/java-11-openjdk-amd64/include -I/usr/lib/jvm/java-11-openjdk-amd64/include/linux -I/usr/local/include -fpic -O3 -fp-model precise -Wall -Wstrict-prototypes -c conftest.c -o conftest.o
icx -shared -L/lustre7/software/intel_ubuntu/oneapi/compiler/2024.0/lib -o conftest.so conftest.o -L/usr/lib/jvm/java-11-openjdk-amd64/lib/server -ljvm
make[2]: Leaving directory '/tmp/Rjavareconf.5WK0Ji'
JAVA_HOME : /usr/lib/jvm/java-11-openjdk-amd64
Java library path: $(JAVA_HOME)/lib/server
JNI cpp flags : -I$(JAVA_HOME)/include -I$(JAVA_HOME)/include/linux
JNI linker flags : -L$(JAVA_HOME)/lib/server -ljvm
Updating Java configuration in /home/yxxxx/R2/R-4.3.2
Done.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2'
Ensure no errors occur during the build process.
Then perform make install:
yxxxx@at138:~/R2/R-4.3.2$ make install
mkdir -p -- /home/yxxxx/R-Intel/lib/R
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/m4'
make[1]: Nothing to be done for 'install'.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/m4'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/tools'
make[1]: Nothing to be done for 'install'.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/tools'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/doc'
installing doc ...
(省略)
icx -I. -I../../src/include -I../../src/include -I/usr/local/include -DHAVE_CONFIG_H -O3 -fp-model precise -Wall -Wstrict-prototypes -L/lustre7/software/intel_ubuntu/oneapi/compiler/2024.0/lib -DR_HOME='"/home/yxxxx/R-Intel/lib/R"' \
-o Rscript ./Rscript.c
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/unix'
(省略)
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/modules'
make[2]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/library'
mkdir -p -- /home/yxxxx/R-Intel/lib/R/library
installing packages ...
building HTML index ...
make[2]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src/library'
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/src'
make[1]: Entering directory '/lustre7/home/yxxxx/R2/R-4.3.2/tests'
make[1]: Nothing to be done for 'install'.
make[1]: Leaving directory '/lustre7/home/yxxxx/R2/R-4.3.2/tests'
Verify that R is installed and starts up correctly:
yxxxx@at138:~/R-Intel/bin$ ls -l
total 100
-rwxr-xr-x 1 yxxxx co-xxx 9135 Mar 16 12:28 R
-rwxr-xr-x 1 yxxxx co-xxx 86192 Mar 16 12:28 Rscript
yxxxx@at138:~/R-Intel/bin$ ./R
R version 4.3.2 (2023-10-31) -- "Eye Holes"
Copyright (C) 2023 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
>
Benchmark results are provided for reference:
yxxxx@at139:~/R-Intel/bin$ cat R-benchmark-25.R | ./R --slave
Loading required package: Matrix
Loading required package: SuppDists
Warning message:
In library(package, lib.loc = lib.loc, character.only = TRUE, logical.return = TRUE, :
there is no package called ‘SuppDists’
Warning messages:
1: In remove("a", "b") : object 'a' not found
2: In remove("a", "b") : object 'b' not found
R Benchmark 2.5
===============
Number of times each test is run__________________________: 3
I. Matrix calculation
---------------------
Creation, transp., deformation of a 2500x2500 matrix (sec): 0.542333333333333
2400x2400 normal distributed random matrix ^1000____ (sec): 0.497333333333333
Sorting of 7,000,000 random values__________________ (sec): 0.790666666666666
2800x2800 cross-product matrix (b = a' * a)_________ (sec): 0.788333333333333
Linear regr. over a 3000x3000 matrix (c = a \ b')___ (sec): 0.379999999999999
--------------------------------------------
Trimmed geom. mean (2 extremes eliminated): 0.596862901307099
II. Matrix functions
--------------------
FFT over 2,400,000 random values____________________ (sec): 0.177999999999998
Eigenvalues of a 640x640 random matrix______________ (sec): 0.215
Determinant of a 2500x2500 random matrix____________ (sec): 0.317
Cholesky decomposition of a 3000x3000 matrix________ (sec): 0.270666666666667
Inverse of a 1600x1600 random matrix________________ (sec): 0.282
--------------------------------------------
Trimmed geom. mean (2 extremes eliminated): 0.254121131799156
III. Programmation
------------------
3,500,000 Fibonacci numbers calculation (vector calc)(sec): 0.259333333333334
Creation of a 3000x3000 Hilbert matrix (matrix calc) (sec): 0.199666666666667
Grand common divisors of 400,000 pairs (recursion)__ (sec): 0.286000000000001
Creation of a 500x500 Toeplitz matrix (loops)_______ (sec): 0.0423333333333341
Escoufier's method on a 45x45 matrix (mixed)________ (sec): 0.218000000000004
--------------------------------------------
Trimmed geom. mean (2 extremes eliminated): 0.224322826166267
Total time for all 15 tests_________________________ (sec): 5.26666666666667
Overall mean (sum of I, II and III trimmed means/3)_ (sec): 0.324038250064507
--- End of test ---
The results show that R without MKL is faster than R with MKL but slower than R operating in multi-thread mode. However, since only one CPU core is used, it is efficient from a resource utilization perspective. It could be viable for non-large-scale problems.